The Chemistry of Unicellular Green Algae (Chlamydomonas reinhardtii)
Ai-Lan Ji-Eun Nguyen, Tesnim Obey, Anders Schwarz, Jackson Yu
Abstract
Chlamydomonas reinhardtii, a unicellular green alga, possesses a sophisticated chemical repertoire that enhances its adaptability and interaction with its environment. This paper delves into the alga’s chemical composition and biological mechanisms, revealing how they contribute to its survival and functional versatility, as well as drawing from the alga’s ingenious natural design solutions to inspire a diverse audience. The alga’s adeptness at photosynthesis and phototaxis is facilitated by specialised structures such as chloroplasts and the eyespot, while Ca2+ signalling plays a crucial role in flagellar movement and cellular response to environmental stimuli. The organism’s remarkable adaptability is further exemplified by its ability to modulate metabolic pathways in response to light availability, enabling it to inhabit diverse ecological niches. This adaptability not only underscores Chlamydomonas reinhardtii‘s ecological success but also positions it as a promising candidate for research and applications in biofuel production, environmental science, and biotechnology.
Introduction
In the diverse world of eukaryotic microorganisms, Chlamydomonas reinhardtii stands out as a model organism that offers profound insights into cellular biology, with implications that span from ecological understanding to biotechnological advancement. This unicellular green alga, equipped with a pair of flagella and a photosynthetic apparatus, has become a cornerstone in biological research, providing a window into the evolutionary past of multicellular plants and animals. The present paper seeks to dissect the multifaceted aspects of Chlamydomonas reinhardtii‘s biology, emphasizing its chemical and physiological mechanisms that confer survival advantages and environmental adaptability.
Photosynthesis and respiration of Chlamydomonas reinhardtii
The unicellular green algae Chlamydomonas reinhardtii can undergo both photosynthesis and respiration, similar to more complex organisms such as plants. Due to their small size(around 10 µm long and 3 µm wide) and unicellular nature, however, the photosynthetic and respiratory processes are performed on a much smaller scale (Rochaix, 2013). Depending on environmental conditions, Chlamydomonas reinhardtii can adapt to switch between performing photosynthesis and respiration to fulfil its energy needs. In the presence of light, it mainly performs photosynthesis (producing energy and organic molecules), while it relies more on respiration to fulfil its energy needs in the absence of light. Both photosynthesis and respiration require a series of chemical reactions.
Photosynthesis in chlamydomonas
Photosynthesis is a biochemical process, involving the conversion of H2O and CO2 into oxygen and glucose molecules, which can be reserved and later broken down to provide the photosynthetic organism with necessary energy in the form of ATP (adenosine triphosphate) (Johnson & Alric, 2013). Typical photosynthesis can be broken down into “light” (light-dependent) and “dark” (light-independent) reactions; “light” reactions involve the absorption of light energy to split into oxygen, protons, and electrons, while “dark” reactions use these proton and electron products from the “light” reactions to reduce CO2 into a starch (Johnson, 2016).
Photosynthetic algae, such as Chlamydomonas, use light energy to drive a series of chemical and redox reactions, in order to regulate atmospheric carbon in the form of CO2 (Johnson & Alric, 2013). As with other algae, each Chlamydomonas cell possesses one chloroplast and multiple (in the range of 10-100) mitochondria (Maul et al., 2002). The primary site of photosynthesis is the chloroplast, a plastid (double membrane-bound organelle) containing the thylakoids and stroma (Maul et al., 2002). Chloroplasts also contain chlorophyll (green pigment) molecules, and other pigments, which absorb the light energy that enters the Chlamydomonas cell body via its eyespot, which behaves as a light-focusing lens.
The main components of photosynthesis in Chlamydomonas reinhardtii are light absorption (involving photosynthetic pigments), photosynthetic membranes, light-dependent reactions, ATP and NADPH production, and the Calvin cycle. The electron transport chain (ETC) is an essential component of the light-dependent reactions involved in photosynthesis, as it is the main player in the conversion of light energy into chemical energy (in the form of ATP and NADPH) (Johnson & Alric, 2013). These energy-rich molecules are essential, as they are later used in the carbon fixation cycle (also known as the Calvin cycle), the stage in photosynthesis that follows the ETC (Yang et al., 2017). In photosynthesis, the ETC takes place within the thylakoid membranes (located in the chloroplasts); in particular, the ETC occurs within photosystem I (PSI) and photosystem II (PSII) (Johnson, 2016). The main purpose of the ETC (in photosynthesis) is to transfer electrons from H2O to NADP+ (Nicotinamide Adenine Dinucleotide Phosphate), which then produces NADPH, a process that is vital for the capture and storage of light energy absorbed from sunlight (Forti, 2008).
There are multiple steps associated with the ETC, beginning with PSII (a protein-pigment complex embedded within the thylakoid membrane), where light energy is stored by chlorophyll molecules and other photosynthetic pigments; this results in the excitation of electrons (Johnson, 2016). The excited electrons pass through a series of various protein complexes within the thylakoid membrane and release energy, which is utilized to pump H+ protons into the thylakoid space, thus creating a proton gradient (Johnson, 2016). Electrons in PSII are then transferred to PSI via plastoquinone, a mobile electron carrier (Johnson & Alric, 2013). Electrons are re-energized in PSI after undergoing light absorption (via chlorophyll molecules) and are transferred to ferredoxin (Fd), another mobile electron carrier (Johnson & Alric, 2013).
The proton gradient created in the thylakoid space during the ETC is also an important byproduct of the ETC as it stores potential energy, which is used by ATP synthase, which drives the synthesis of ATP from ADP and inorganic phosphate (see Figure 1) (Minagawa & Tokutsu, 2015). As the ETC is characterized by the controlled flow of electrons, this aids in preventing the formation of potentially harmful reactive oxygen species, such as superoxide radicals, which could damage the physical components of photosynthesis (Gollan et al., 2017).
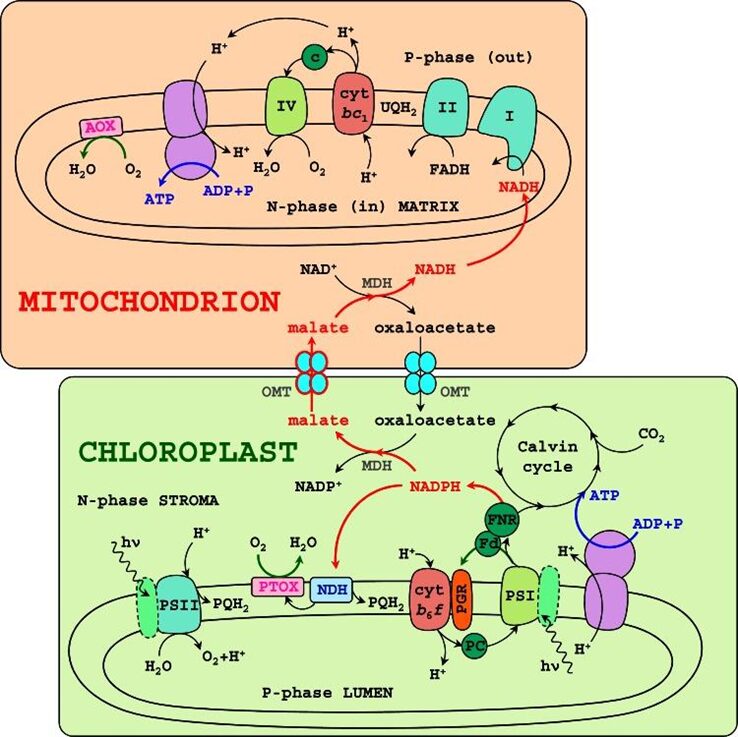
Fig. 1. Diagram of energetic organelles, the mitochondria, and chloroplast, illustrating interactions between photosynthesis and respiration in Chlamydomonas reinhardtii. Black arrows signify proton and electron transfer, and blue arrows show ATP synthesis. Notice how NADPH and ATP are sent to the Calvin cycle via light reactions and the ETC in the chloroplast. Red arrows signify alternative pathways of electron transfers (Johnson & Alric, 2013).
Respiration in Chlamydomonas reinhardtii
Respiration refers to the process through which organisms are able to extract energy from organic compounds (such as starch) to power their metabolic activities (Hill, 2014). Chlamydomonas reinhardtii undergoes respiration to produce the necessary energy for growth and maintenance (Raven & Beardall, 2003). The main components of respiration in Chlamydomonas reinhardtii include glycolysis, mitochondrial respiration, the electron transport chain, ATP synthesis, and CO2 production (Hill, 2014).
The first step of respiration is glycolysis, the process in which glucose molecules are broken down into pyruvate, which takes place in the cytoplasm of the cell (Chen & Gibbs, 1991). This process generates a small amount of ATP and NADPH (Chen & Gibbs, 1991). The pyruvate produced by glycolysis is transported to the mitochondria, where it then enters the citric acid cycle (colloquially referred to as the Krebs cycle) and the ETC (Gérin et al., 2014). These two processes generate much more ATP than glycolysis, through oxidative phosphorylation (Gérin et al., 2014). This ATP is eventually used to power cellular processes such as growth, maintenance, and even cell division (Raven & Beardall, 2003).
TOR kinase and regulating respiration vs photosynthesis
Target-of-Rapamycin (TOR) kinase, involved in maintaining homeostasis of the cell, has been linked to the regulation of photosynthetic chloroplast and respiratory mitochondria functions in Chlamydomonas reinhardtii (Upadhyaya & Rao, 2019). TOR kinase regulates cell growth and metabolic processes in response to various environmental changes, such as the presence (or lack) of light, and by sensing the energy levels of the cell (McCready et al., 2020). As photosynthetic organisms rely on their chloroplasts and mitochondria for energy metabolism, communication between organelles is essential to the proper occurrence of the series of chemical reactions involved in photosynthesis (Upadhyaya & Rao, 2019). One experiment demonstrated how the inhibition of TOR kinase in a Chlamydomonas cell can impair the maintenance of certain mechanisms associated with PSII and can also inhibit efficient transitions between PSII and PSI (Upadhyaya & Rao, 2019).
Applications in biofuel research
One pursuit in protecting the environment, given the modern fight against climate change, while also potentially maintaining today’s current economy, is the development of new renewable energy sources (Scranton et al., 2015). It so happens that algal biomass, such as lipids and pigments, and metabolites, such as glucose produced from photosynthesis, present a promising alternative source of biofuel (Banerjee et al., 2021). Current biofuels rely mostly on food or oil crops, however, due to their demand for plowable (arable) land, which overall has a negative impact on global food security (Banerjee et al., 2021). Biofuel sourced from algal biomass on the other hand, specifically from Chlamydomonas reinhardtii, do not present this issue as they do not compete for arable land, as they can grow in most conditions (including wastewater), and have relatively quick regenerative times (Banerjee et al., 2021). As such, algae as an alternative source of biofuel is currently a research area being widely explored (Dubini, 2011). To date, research in this field has been mainly on Chlamydomonas biomass as a source of biodiesel, however, it should be noted that other sources of fuel, such as bioethanol (alcohol) and biogas (hydrogen), are possible using algal biomass (see Figure 2) (Scranton et al., 2015).
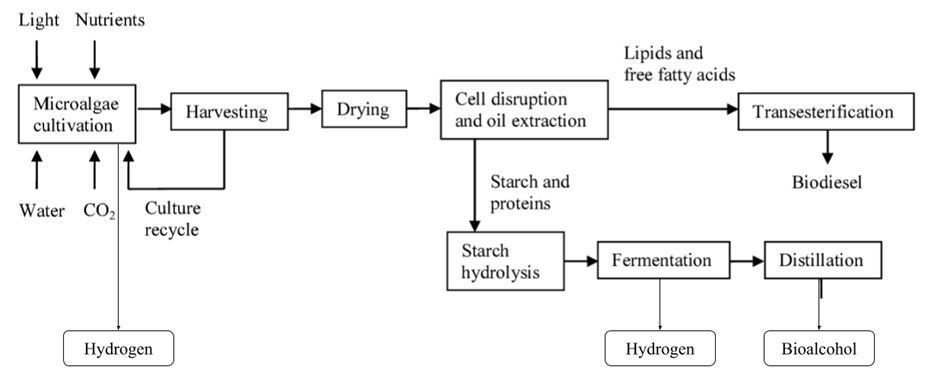
Fig. 2. Production steps of hydrogen, bio-alcohol, and biodiesel from cultivated algal biomass. Biofuel production with algal biomass requires multiple steps, including microalgae cultivation (either under special conditions in bioreactors or under natural conditions in ponds), biomass collection and filtration, chemical or mechanical pretreatment of biomass (such as drying), chemical or thermal conversion of biomass to biofuel, and eventual purification of biofuel products (Voloshin et al., 2016).
Alternative Fertilizer: More and more people worldwide are promoting eco-friendly agricultural practices (Zou et al., 2021). It turns out that algal biomass also has various potential benefits as a biofertilizer. Bio-fertilizers are a pollution-free alternative for increasing soil nutrient content for agricultural purposes (Ammar et al., 2022). As such, biofertilizer derived from algal biomass provides a non-toxic, non-carcinogenic alternative to current inorganic and organic fertilizers, which have historically contributed to the aggregation of harmful heavy metals in soil (Ammar et al., 2022).
Algal biomass, such as biomass sourced from Chlamydomonas, presents many opportunities to meet current global energy needs. However, many improvements still need to be made in order to make this energy source more accessible. With their current production methods, algal biofuels are still not economically viable, and manufacturing is quite energy-consuming (which defeats the purpose of developing an alternative, renewable energy source); this limits its overall commercialization (Banerjee et al., 2021).
Flagella
Flagella, or cilia, are found in many biflagellate unicellular green algae but also other organisms such as in the human lungs. Therefore, understanding the flagellar system is crucial for understanding human health. Luckily, Chlamydomonas’ flagellar system has been researched abundantly, providing a solid base for researchers and for us who want to explore the wonders of unicellular algae.
Flagella composition
The flagella’s backbone is composed of alpha and beta-tubulin – polarised protein molecules- which form a dimer containing a plus end toward the beta-tubulin and a minus end in the alpha-tubulin. The dimers assemble in a spiral, growing towards the positive end. The rate-limiting step is nucleation to create the microtubule seed, which means that reducing the energy necessary to overcome that step reduces the overall energy needed significantly (Conde & Cáceres, 2009). Nucleation involves the assembly of tubulin subunits into a small, stable structure known as a “nucleus” or “seed.” This seed serves as the starting point for microtubule growth. The nucleation process can occur in several ways. For instance, de novo nucleation is the spontaneous formation of a microtubule seed in the absence of pre-existing microtubules (Dominguez, 2010). In cells, this can occur at specific cellular structures or microtubule organising centres (MTOCs), such as the centrosome or basal body. De novo nucleation is crucial for initiating new microtubules in the cell. However, this process is time and energy-consuming, especially for green algae which rely on their flagella for feeding, movement, and adhesion amongst other necessary functions. So, most eukaryotic cells rely on template-driven nucleation which uses pre-existing microtubules as templates for the formation of new microtubules (Stephens & Edds, 1976). In this process, tubulin subunits are added to an existing microtubule, helping extend its length. This mechanism is partly achieved by using a centriole, a grouping of microtubules, as the template, and the complex allowing for nucleation is referred to as the basal body. Unlike the rest of the flagella, or the axoneme, which remains outside of the cell body, basal bodies remain in the cellular body which protects it from extracellular disturbances. The centriole contains γ-tubulin, which eases axoneme microtubule assembly making the nucleation process, the rate-limiting step, less energy-consuming (Silflow et al., 1999). Following the structure of the centriole, two microtubules combine to form a doublet microtubule, and nine of those doublets are formed on the outside as shown in Figure 3.

Fig. 3. Intersection of a 9+2 flagella structure (Mitchell, 2000).
Another doublet also forms in the centre and this composition is referred to as the “9+2” structure. This formation of microtubules gives the flagella its structural integrity, but that alone does not ensure its motility nor its various other characteristics such as adhesion or intraflagellar transport which allow the survival of many biflagellate unicellular green algae and many eukaryotic cells.
In addition to tubulin, the Chlamydomonas’ flagella contain hundreds of protein complexes, many of which are not identified on a molecular level or from a functional point of view (Lechtreck et al., 2009). These proteins form enzymes, motor proteins, and photoproteins in unicellular green algae. Like in several mammalian biological systems, an essential motor protein in flagella is dynein, which needs adenosine triphosphate (ATP) to function (Smith, 2002) (Boesger et al., 2009). On each doublet microtubule exists a row of inner dynein complexes and an outer row as shown in Figure 4.
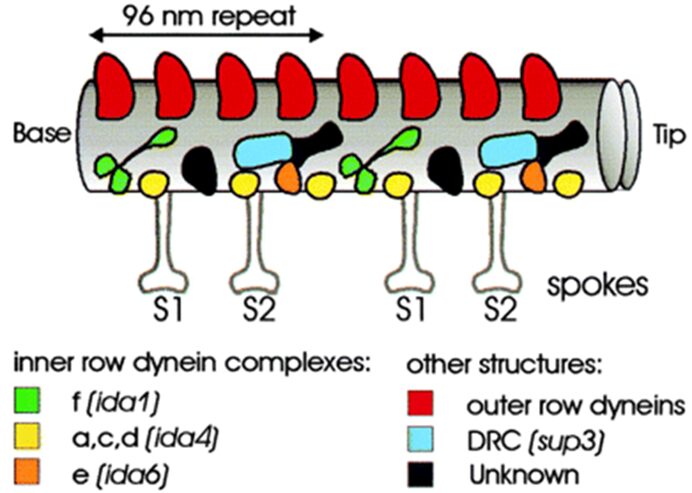
Fig. 4. Diagram of dynein complexes and spokes in motile flagella microtubule (Mitchell, 2000).
In addition to those proteins, proteome analysis– separation, identification, and measurement of the entire protein complement expressed by a genome, cell, or tissue- has revealed the presence of several kinases and protein phosphatases which are enzymes that catalyse phosphorylation and dephosphorylation of molecules. Through a cycle of phosphorylation and dephosphorylation, the dyneins experience continuous conformational change of their protein complex or switch (Ishikawa, 2017). This switching model proposes that the bending motion of motile cilia is driven by a coordinated switching of the states of dynein molecules on one side of the curved cilia compared to the other side. This orchestrated switch in dynein states is believed to create or maintain asymmetry, thereby leading to the bending and movement of cilia (King, 2016). While this model provides insights into the mechanics of ciliary motility, it does not explain the rapid ciliary movement as this process alone would yield slower movement. While the mechanisms allowing for the rapid ciliary movements are still under investigation, a prominent explanation is the involvement of the calcium cation in both the control of the flagellar beating velocity as well as the change of phototaxis. In Chlamydomonas, switching between forward motion using an asymmetric or ciliary beat and backward movement driven by a symmetric sinusoidal waveform is mediated by altering intraciliary Ca2+ (Bessen et al. 1980). When Ca2+ levels increase to 10-5 M, the cilia stop beating briefly and restart with a different waveform. The outer dynein row in the cilia is crucial for this transition. Mutated species lacking that row show an irregular waveform or stop beating when Ca2+ levels rise. The protein LC4 seems heavily involved in these processes. This protein binds to two distinct segments of the γ heavy-chain amino-terminal region and, in the absence of Ca2+, is rigid which maintains the bent orientation of the flagella. However, when the levels of Ca2+ increase, LC4 appears to detach from the basal body which allows the γ-tubulin heavy basal body to adopt conformations not attainable in the absence of the ligand. However, how this allows for the change in beating frequency is still unknown (Bessen et al., 1980).
Another crucial protein complex in flagella motility is the radial spoke. Radial spokes are T-shaped structures found within the axoneme, consisting of a “head” and a “stalk,” with each of these sub-structures made up of numerous protein subunits. The radial spoke contains at least seventeen proteins, with five in the head and twelve in the stalk (Yang et al., 2006). The head is oriented towards the central microtubule pair. Mutated Chlamydomonas lacking these central components have paralysed flagella. However, the exact role of this apparatus in the regulation of the waveform and the bending of the flagella is still under question. Some evidence points to the central pair interacting with radial spokes, which transmits regulatory systems to the inner and outer rows of dynein and potentially the enzymes that regulate phosphorylation. However, the flagella lacking the central apparatus are still photo-responsive (Yokoyama et al., 2004; Loreng & Smith, 2017). The inhibition of the proteins attached to the axoneme not influencing many of the cell’s photo responses suggests that many photoproteins are found in the plasma membrane of the flagella. The plasma membrane potentially contains a pool of surface-exposed (biotinylated) proteins that regulate flagellar activities.
Revolutionary discovery: intraflagellar transport
The maintenance of the flagella is a challenge as they are appendages outside of the cellular body, which is a topic that puzzled scientists. Chlamydomonas led to the discovery of intraflagellar transport in 1997 by Piperno and Mead, which answers how the flagella are able to be maintained, assembled, and even disassembled when needed (Piperno & Mead, 1997).
Intraflagellar transport (IFT) is the bidirectional movement of transporters between the basal body and the tip. The staging area for the IFT proteins and the binding of flagellar proteins (such as dyneins or stokes) is the transitional fibres of unknown composition which run between the basal body centriole and the plasma membrane (Wei et al., 2015). This ensures that the only proteins coming from the basal body towards the tip (this direction is called anterograde IFT) enter the tip which maintains the proper structure and functioning of the flagella. This process is most likely controlled by protein-protein binding regulators such as TPR, commonly found in flagellated eukaryotic cells. Anterograde IFT is driven by the motor protein kinesin (Cole, 2003). This protein seems to be walking along the microtubule as it is phosphorylated and dephosphorylated, akin to dynein as shown in Figure 5. Dynein however plays a major part in retrograde IFT, which is the transport from the flagella appendage towards the basal body as shown in Figure 6. In Chlamydomonas, 16 proteins involved in IFT have been identified and they most likely play a crucial part in signal transduction on top of flagellar maintenance (Satarić et al., 2023).


Fig. 5. Similarities between the phosphorylation-dependent movement of kinesin along microtubules towards the positive end (1) and the phosphorylation-dependent movement of dynein through the power stroke towards the negative end along the microtubule (adapted from Rossien et al. 2015) (2).

Fig. 6. Intraflagellar transport model:Anterograde and Retrograde Movement via Kinesin-II and Cytoplasmic Dynein 1b Along Outer Doublet Microtubules.
Chlamydomonas’ unique solution
Chlamydomonas developed a unique strategy pertaining to the maintenance of its flagella. When one flagellum gets cut off, it will start building the new flagellum while breaking down the longer flagellum. This is done until both are at the same length and only then would both be rebuilt at the same rate until the original length is restored, which is typically between 10-14 micrometres (Mitchell, 2000). This makes sense as the movement of the Chlamydomonas relies on the interplay and synchronisation of the flagella beating. This can be paralleled to rowing a boat. If both paddles are producing the same force, then the boat will move in a straight line. But, if a paddle is missing, the boat will only move in a circular motion, which is not effective. Furthermore, the presence of the appropriate microtubule is not guaranteed, so the cell likely uses the disassembled proteins to reconstruct the shortened flagella as the process has been shown to not be linked to protein synthesis. However, the building of both flagella leads to a spike in tubulin and axoneme-related proteins mRNA in the intracellular matrix.
The biflagellated unicellular green algae Chlamydomonas provides insight into flagellar structures and operation. Flagella are not solely vital for the existence and operation of numerous unicellular organisms like Chlamydomonas, but they also hold a notable significance in more sophisticated organisms, including mammalian lungs and other cellular mechanisms. Comprehending the complex flagellar system is imperative for acquiring knowledge about human well-being and diverse biological functionalities.
Calcium concentration and signaling
Control of ion concentration within the cell is an essential aspect of maintaining homeostasis for all cells. The regulation of cytosolic calcium ion concentration [Ca2+] in particular is a necessity given the cytotoxicity of this ion that could cause clumping of proteins and nucleic acid, as well as precipitation of phosphates from prolonged exposure to increased concentrations (Ballotari & Pivato, 2021). Therefore, the first unicellular cells possessed particular transport systems to maintain Ca2+ concentration at a desired level of about 10,000 to 20,000 times lower than the extracellular concentration (Ballotari & Pivato, 2021). The existence of a concentration gradient at such a magnitude produced the evolutionary environment for the development of Ca2+ signaling system.
Chlamydomonas is one such organism that utilizes this system, with calcium Ca2+ ion-dependent signaling methods being central to many responses in Chlamydomonas, including flagella function, photosynthesis, and stress signaling (Wheeler, 2017). Chlamydomonas reinhardtii has been found to possess ‘animal-like’ channels for calcium diffusion down an electrochemical gradient like transient receptor potential channels, voltage-gated cation channels, and inositol trisphosphate receptors, all of which are distinctly absent from plant genomes (Ballotari & Pivato, 2021). These channels in addition to standard plant components for Ca2+ channels, with Ca2+ transporters to move against the concentration gradient using ATP, allow Chlamydomonas to effectively mediate Ca2+ concentration, opening the door to the use of this ion as a form of signaling in response to various stimuli, with some examples following.
In Chlamydomonas, the eyespot is an apparatus capable of detecting the direction and intensity of light (Ballotari & Pivato, 2021), allowing the organism to decide when and how to perform phototaxis. The morphology of this organelle has been shown through electron microscopy to consist of a distinct region containing carotenoid granules that are hexagonally packed, functioning as a wave interference reflector (Kato et al, 2019). The signalling behind phototaxis has been shown to be mediated by plasma membrane-located photoreceptors channelrhodopsin (ChR), light-gated channels capable of allowing rapid Ca2+ influx current from the extracellular region towards the eyespot region (Ballotari & Pivato, 2021). A Ca2+ feedback loop therefore regulates ChR function, and it has been shown that the phosphorylation involved in energy exchange at ChR’s is an important aspect of the phototactic sensitivity present in Chlamydomonas reinhardtii (Böhm et al., 2019). Hence, a flash stimulation on Chlamydomonas has been observed to provoke an influx of calcium current into the eyespot and flagella. This photocurrent has even been suggested to be the trigger for all behavioral light responses of the cell (Holland et al, 1997). With several Ca2+ binding proteins and transporters having been identified on subcellular fractions of intact eyespots on Chlamydomonas reinhardtii, there is a wealth of evidence to suggest calcium ions play a crucial role in signal transmission and light sensing processes, thereby mediating the corresponding flagella beating as part of the following phototactic response (Ballotari & Pivato, 2021).

Fig. 7. Numerous organelles and apparatus within Chlamydomonas are linked to calcium Ca2+ ions, that are capable of influx/efflux through all the indicated regions. While the exact function of these relationships is more clearly defined aspects like the Flagella and eyespot, others are less understood such as the mitochondrial role in intracellular Ca2+ dynamics, despite these organelles’ ability to rapidly accumulate this ion (Ballotari & Pivato, 2021).
As a model organism for flagella function and structure (Ballotari & Pivato, 2021), studies have concluded that calcium regulates flagellar motility through the control of dynein-driven microtubule sliding through a calcium signalling pathway. Located on the outside of the body, the axoneme is a microtubule-built structure forming the core of the flagellum, and it contains several highly conserved calcium-binding proteins (Smith, 2002) that transduce the presence of calcium to dynein motor proteins that drive flagellar movement through microtubule sliding. Indeed, dynein activity in Chlamydomonas increases linearly with increasing concentrations of calcium. This suggests the effectiveness of the calcium signalling system in provoking flagellar beating, with the concentration of calcium effectively moderating the flagella through interaction with calcium-sensing proteins. With the ability to increase calcium concentrations without expending energy considering the aforementioned significant concentration gradient difference between the extra and intracellular environments, calcium signalling effectively allows Chlamydomonas to control flagellar beating, which is itself a response to phototaxis which is provoked by the eyespot which is additionally moderated through calcium concentration levels.
Additionally, calcium signalling has been shown to be critical in activating responses to osmotic stress provoked by salinity. Exposure to salt stress induced a measured elevation in calcium originating in the cell that spread as a “calcium wave” from the centre of the cell to the surrounding cytosol. Exposure to hypo-osmotic conditions was also found to induce calcium elevations in the flagella that occurred independently from those in the cytosol (Bickerton et al, 2016), demonstrating the diverse breadth of signalling that calcium ions are utilized for by Chlamydomonas.
While Ca2+ signalling has been shown to influence various aspects of Chlamydomonas as shown in Figure 7 with some cases well documented, the scientific understanding of Ca2+ signalling is considered to be in its infancy with little known about the detailed mechanisms through which this signalling operates (Ballotari & Pivato, 2021).
Oxidative stress response and autophagy
Another important aspect of maintaining homeostasis in cells is avoiding oxidative stress: a phenomenon that emerges from an imbalance between the formation of free radicals and the ability of the cell to clear them. An example would be a reactive oxygen species (ROS) produced as a product of metabolism by an aerobic photosynthetic organism like Chlamydomonas, which could damage the cell membrane through lipid peroxidation and could strengthen toxins (Ma et al, 2020). Therefore, to combat the inevitable threats posed by an abundance of ROS, Chlamydomonas cells have an antioxidant system designed to cope with this oxidative stress.
Enzymes in Chlamydomonas reinhardtii serve generally as the most immediate responders to oxidative stress. One such enzyme is the glutathione peroxidase enzyme (GPX), which catalyses the detoxification of organic peroxides with the protein thioredoxin as an electron donor (Ma et al, 2020). Studies have highlighted the crucial presence of GPX by removing this enzyme from Chlamydomonas and subjecting the cells to oxidative stress, and those lacking this enzyme suffered ROS levels that were three times higher (Ma et al, 2020). Ultimately, Chlamydomonas can effectively moderate the expression and activity of these enzymes to ensure an efficient detoxification process.
Autophagy: a critical form of recycling system for the cell in which damaged or obsolete cytoplasmic material is degraded or recycled through a set of pathways involving the lysosome organelle, which acts as a digestive system. Derived from the greek expression for “self-eating”, autophagy can be especially observed in cells experiencing starvation.
However, when under severe oxidative stress, Chlamydomonas will accelerate their citric acid cycle to produce more ATP, as well as down-regulating iron transporters to avoid protection from damage to heavier iron atoms by radicals (Ma et al, 2020). Additionally, the process of autophagy can be induced in which cytoplasmic components are enclosed by a vesicle called an autophagosome transports the enclosed components to a vacuole or lysosome for the degradation and potential conservation of damaged or toxic components. This has been shown to happen for Chlamydomonas cells treated with hydrogen peroxide or methyl viologen radicals, with this oxidative stress triggering the autophagy process (Martín et al, 2014).
In other words, autophagy has been observed to occur in Chlamydomonas as a stress response to a variety of stimuli. It has even been found that under proteotoxic stress from misfolded or damaged proteins, Chlamydomonas can go as far as to perform autophagy on chloroplast proteins (Li et al, 2021), while another study inducing autophagy under high light conditions found that under lethal high-intensity illumination, the autophagic pathway in Chlamydomonas is regulated by nitrogen oxide (Kuo et al, 2020). Also, Chlamydomonas can employ sophisticated transcriptional regulatory mechanisms to control oxidative stress, by upregulating the expression of genes encoding antioxidant enzymes to ensure a rapid and efficient response to increased ROS levels (Ma et al, 2020).
Conclusion
Throughout this exploration of Chlamydomonas reinhardtii, we have witnessed the alga’s remarkable biological design solutions that enable it to thrive in a diverse range of environmental conditions. The alga’s photosynthetic efficiency, governed by the interplay between its chloroplasts and eyespot, exemplifies a highly refined system evolved to maximise energy capture from light. This system is not a mere consequence of chance, but a sophisticated adaptation honed by millions of years of evolutionary pressure.
The flagellar apparatus of Chlamydomonas reinhardtii, regulated by intricate Ca2+ signalling pathways, is another testament to the organism’s evolutionary ingenuity. These flagella are not only locomotive organelles but also serve as sensory and signalling devices that allow the alga to respond to its environment with precision and agility. The design of these flagella, with their complex assembly of microtubules and motor proteins, is a marvel of nature’s engineering.
Moreover, the metabolic versatility of Chlamydomonas reinhardtii, particularly its ability to modulate its pathways in response to light availability, underscores a design solution that is both elegant and practical. This adaptability ensures the alga’s survival across various habitats and light regimes, making it a resilient and persistent component of the ecosystem.
In the realm of biotechnology, the design solutions inherent in Chlamydomonas reinhardtii offer promising avenues for innovation. The alga’s metabolic pathways, especially those related to lipid production, present opportunities for biofuel development, potentially contributing to a more sustainable energy future. The algae’s ability to adapt its metabolism has significant implications for environmental science and biotechnological applications, where its mechanisms can be harnessed and optimised for human benefit.
Chlamydomonas reinhardtii serves as a living blueprint of nature’s problem-solving prowess. The insights gained from studying this alga’s design solutions not only enhance our understanding of biological systems but also inspire us to emulate these natural strategies in addressing contemporary challenges. As we continue to unravel the complexities of Chlamydomonas reinhardtii, we pave the way for innovative approaches that bridge the gap between biological understanding and technological advancement.
References
Ammar, E. E., Aioub, A. A. A., Elesawy, A. E., Karkour, A. M., Mouhamed, M. S., Amer, A. A., & El-Shershaby, N. A. (2022). Algae as Bio-fertilizers: Between current situation and future prospective. Saudi Journal of Biological Sciences, 29(5), 3083-3096. https://doi.org/https://doi.org/10.1016/j.sjbs.2022.03.020
Banerjee, S., Ray, A., & Das, D. (2021). Optimization of Chlamydomonas reinhardtii cultivation with simultaneous CO2 sequestration and biofuels production in a biorefinery framework. Science of The Total Environment, 762, 143080. https://doi.org/https://doi.org/10.1016/j.scitotenv.2020.143080
Bessen, M., Fay, R. B., & Witman, G. B. (1980). Calcium control of waveform in isolated flagellar axonemes of Chlamydomonas. J Cell Biol, 86(2), 446-455. https://doi.org/10.1083/jcb.86.2.446
Bickerton, P., Sello, S., Brownlee, C., Pittman, J. K., & Wheeler, G. L. (2016). Spatial and temporal specificity of Ca2+ signalling in Chlamydomonas reinhardtii in response to osmotic stress. New Phytologist, 212(4), 920-933. https://doi.org/https://doi.org/10.1111/nph.14128
Boesger, J., Wagner, V., Weisheit, W., & Mittag, M. (2009). Analysis of flagellar phosphoproteins from Chlamydomonas reinhardtii. Eukaryot Cell, 8(7), 922-932. https://doi.org/10.1128/ec.00067-09
Böhm, M., Boness, D., Fantisch, E., Erhard, H., Frauenholz, J., Kowalzyk, Z., Marcinkowski, N., Kateriya, S., Hegemann, P., & Kreimer, G. (2019). Channelrhodopsin-1 Phosphorylation Changes with Phototactic Behavior and Responds to Physiological Stimuli in Chlamydomonas. Plant Cell, 31(4), 886-910. https://doi.org/10.1105/tpc.18.00936
Chen, C., & Gibbs, M. (1991). Glucose Respiration in the Intact Chloroplast of Chlamydomonas reinhardtii. Plant Physiol, 95(1), 82-87. https://doi.org/10.1104/pp.95.1.82
Cole, D. G. (2003). Intraflagellar transport in the unicellular green alga, Chlamydomonas reinhardtii. Protist, 154(2), 181-191. https://proxy.library.mcgill.ca/login?url=https://www.proquest.com/scholarly-journals/intraflagellar-transport-unicellular-green-alga/docview/207944095/se-2
Cole, D. G. (2003). The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic, 4(7), 435-442. https://doi.org/10.1034/j.1600-0854.2003.t01-1-00103.x
Conde, C., & Cáceres, A. (2009). Microtubule assembly, organization and dynamics in axons and dendrites. Nature Reviews Neuroscience, 10(5), 319-332. https://doi.org/10.1038/nrn2631
Dominguez, R. (2010). Structural insights into de novo actin polymerization. Curr Opin Struct Biol, 20(2), 217-225. https://doi.org/10.1016/j.sbi.2009.12.012
Dragone, G., Fernandes, B. D., Vicente, A. A., & Teixeira, J. A. C. (2010). Third generation biofuels from microalgae.
Dubini, A. (2011). Green energy from green algae: Biofuel production from Chlamydomonas reinhardtii. The Biochemist, 33(2), 20-23. https://doi.org/10.1042/bio03302020
Forti, G. (2008). The role of respiration in the activation of photosynthesis upon illumination of dark adapted Chlamydomonas reinhardtii. Biochimica et Biophysica Acta (BBA) – Bioenergetics, 1777(11), 1449-1454. https://doi.org/https://doi.org/10.1016/j.bbabio.2008.08.011
Gérin, S., Mathy, G., & Franck, F. (2014). Modeling the dependence of respiration and photosynthesis upon light, acetate, carbon dioxide, nitrate and ammonium in Chlamydomonas reinhardtiiusing design of experiments and multiple regression. BMC Systems Biology, 8(1), 96. https://doi.org/10.1186/s12918-014-0096-0
Gollan, P. J., Lima-Melo, Y., Tiwari, A., Tikkanen, M., & Aro, E. M. (2017). Interaction between photosynthetic electron transport and chloroplast sinks triggers protection and signalling important for plant productivity. Philos Trans R Soc Lond B Biol Sci, 372(1730). https://doi.org/10.1098/rstb.2016.0390
Hill, G. E. (2014). Cellular Respiration: The Nexus of Stress, Condition, and Ornamentation. Integrative and Comparative Biology, 54(4), 645-657. https://doi.org/10.1093/icb/icu029
Holland, E. M., Harz, H., Uhl, R., & Hegemann, P. (1997). Control of phobic behavioral responses by rhodopsin-induced photocurrents in Chlamydomonas. Biophysical journal, 73(3), 1395-1401. https://doi.org/https://doi.org/10.1016/S0006-3495(97)78171-2
Huang, B., Ramanis, Z., Dutcher, S. K., & Luck, D. J. (1982). Uniflagellar mutants of Chlamydomonas: evidence for the role of basal bodies in transmission of positional information. Cell, 29(3), 745-753. https://doi.org/10.1016/0092-8674(82)90436-6
Ishikawa, T. (2017). Axoneme Structure from Motile Cilia. Cold Spring Harb Perspect Biol, 9(1). https://doi.org/10.1101/cshperspect.a028076
Johnson, M. P. (2016). Photosynthesis. Essays Biochem, 60(3), 255-273. https://doi.org/10.1042/ebc20160016
Johnson, X., & Alric, J. (2013). Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot Cell, 12(6), 776-793. https://doi.org/10.1128/ec.00318-12
Kato, S., Ozasa, K., Maeda, M., Tanno, Y., Tamaki, S., Higuchi-Takeuchi, M., Numata, K., Kodama, Y., Sato, M., Toyooka, K., & Shinomura, T. (2020). Carotenoids in the eyespot apparatus are required for triggering phototaxis in Euglena gracilis. Plant J, 101(5), 1091-1102. https://doi.org/10.1111/tpj.14576
King, S. M. (2016). Axonemal Dynein Arms. Cold Spring Harb Perspect Biol, 8(11). https://doi.org/10.1101/cshperspect.a028100
Kuo, E. Y., Chang, H.-L., Lin, S.-T., & Lee, T.-M. (2020). High Light-Induced Nitric Oxide Production Induces Autophagy and Cell Death in Chlamydomonas reinhardtii [Original Research]. Frontiers in Plant Science, 11. https://doi.org/10.3389/fpls.2020.00772
Lechtreck, K. F., Luro, S., Awata, J., & Witman, G. B. (2009). HA-tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil Cytoskeleton, 66(8), 469-482. https://doi.org/10.1002/cm.20369
Li, N., Nakamura, S., Ramundo, S., Nishimura, Y., Hagihara, S., & Izumi, M. (2021). Chloroplast proteotoxic stress-induced autophagy is involved in the degradation of chloroplast proteins in Chlamydomonas reinhardtii. Plant and Cell Physiology, 62(4), e1-e31. https://doi.org/10.1093/pcp/pcab029
Lodish, H. F. (2008). Molecular cell biology. Macmillan.
Loreng, T. D., & Smith, E. F. (2017). The Central Apparatus of Cilia and Eukaryotic Flagella. Cold Spring Harb Perspect Biol, 9(2). https://doi.org/10.1101/cshperspect.a028118
Luck, D., Piperno, G., Ramanis, Z., & Huang, B. (1977). Flagellar mutants of Chlamydomonas: Studies of radial spoke-defective strains by dikaryon and revertant analysis. Proceedings of the National Academy of Sciences, 74(8), 3456-3460. https://doi.org/10.1073/pnas.74.8.3456
Ma, X., Zhang, B., Miao, R., Deng, X., Duan, Y., Cheng, Y., Zhang, W., Shi, M., Huang, K., & Xia, X. Q. (2020). Transcriptomic and Physiological Responses to Oxidative Stress in a Chlamydomonas reinhardtii Glutathione Peroxidase Mutant. Genes (Basel), 11(4). https://doi.org/10.3390/genes11040463
Mano, L. Y., Torres, A. M., Morales, A. G., Cruz, C. C. P., Cardoso, F. H., Alves, S. H., Faria, C. O., Lanzillotti, R., Cerceau, R., da Costa, R. M. E. M., Figueiredo, K., & Werneck, V. M. B. (2023). Machine Learning Applied to COVID-19: A Review of the Initial Pandemic Period [Review]. International Journal of Computational Intelligence Systems, 16(1), Article 73. https://doi.org/10.1007/s44196-023-00236-3
Maul, J. E., Lilly, J. W., Cui, L., dePamphilis, C. W., Miller, W., Harris, E. H., & Stern, D. B. (2002). The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell, 14(11), 2659-2679. https://doi.org/10.1105/tpc.006155
McCready, K., Spencer, V., & Kim, M. (2020). The Importance of TOR Kinase in Plant Development [Mini Review]. Frontiers in Plant Science, 11. https://doi.org/10.3389/fpls.2020.00016
McFadden, G. I., Schulze, D., Surek, B., Salisbury, J. L., & Melkonian, M. (1987). Basal body reorientation mediated by a Ca2+-modulated contractile protein. J Cell Biol, 105(2), 903-912. https://doi.org/10.1083/jcb.105.2.903
Minagawa, J., & Tokutsu, R. (2015). Dynamic regulation of photosynthesis in Chlamydomonas reinhardtii. The Plant Journal, 82(3), 413-428. https://doi.org/https://doi.org/10.1111/tpj.12805
Mitchell, D. (2000). Chlamydomonas flagella. Journal of Phycology, 36, 261-273. https://doi.org/10.1046/j.1529-8817.2000.99218.x
Mitchell, D. R., & Sale, W. S. (1999). Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J Cell Biol, 144(2), 293-304. https://doi.org/10.1083/jcb.144.2.293
Omoto, C. K., & Brokaw, C. J. (1985). Bending patterns of Chlamydomonas flagella: II. Calcium effects on reactivated Chlamydomonas flagella. Cell Motil, 5(1), 53-60. https://doi.org/10.1002/cm.970050105
Pérez-Martín, M., Pérez-Pérez, M. E., Lemaire, S. D., & Crespo, J. L. (2014). Oxidative Stress Contributes to Autophagy Induction in Response to Endoplasmic Reticulum Stress in Chlamydomonas reinhardtii Plant Physiology, 166(2), 997-1008. https://doi.org/10.1104/pp.114.243659
Piperno, G., & Mead, K. (1997). Transport of a novel complex in the cytoplasmic matrix of <i>Chlamydomonas </i>flagella. Proceedings of the National Academy of Sciences, 94(9), 4457-4462. https://doi.org/doi:10.1073/pnas.94.9.4457
Pivato, M., & Ballottari, M. (2021). Chlamydomonas reinhardtii cellular compartments and their contribution to intracellular calcium signalling. J Exp Bot, 72(15), 5312-5335. https://doi.org/10.1093/jxb/erab212
Raven, J. A., & Beardall, J. (2003). Carbohydrate Metabolism and Respiration in Algae. In A. W. D. Larkum, S. E. Douglas, & J. A. Raven (Eds.), Photosynthesis in Algae (pp. 205-224). Springer Netherlands. https://doi.org/10.1007/978-94-007-1038-2_10
Raven, J. A., & Beardall, J. (2003). Carbohydrate Metabolism and Respiration in Algae. In A. W. D. Larkum, S. E. Douglas, & J. A. Raven (Eds.), Photosynthesis in Algae (pp. 205-224). Springer Netherlands. https://doi.org/10.1007/978-94-007-1038-2_10
Rochaix, J. D. (2013). Chlamydomonas reinhardtii. In S. Maloy & K. Hughes (Eds.), Brenner’s Encyclopedia of Genetics (Second Edition) (pp. 521-524). Academic Press. https://doi.org/https://doi.org/10.1016/B978-0-12-374984-0.00230-8
Roossien, Douglas & Miller, Kyle & Gallo, Gianluca. (2015). Ciliobrevins as Tools for Studying Dynein Motor Function. Frontiers in cellular neuroscience. 9. 252. 10.3389/fncel.2015.00252.
Rosenbaum, J. L., Moulder, J. E., & Ringo, D. L. (1969). Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol, 41(2), 600-619. https://doi.org/10.1083/jcb.41.2.600
Satarić, M. V., Nemeš, T., & Zdravković, S. (2023). Calcium messages in flagella are faster than messenger particles. Biosystems, 232, 105003. https://doi.org/https://doi.org/10.1016/j.biosystems.2023.105003
Scranton, M. A., Ostrand, J. T., Fields, F. J., & Mayfield, S. P. (2015). Chlamydomonas as a model for biofuels and bio-products production. The Plant Journal, 82(3), 523-531. https://doi.org/https://doi.org/10.1111/tpj.12780
Silflow, C., Liu, B., LaVoie, M., Richardson, E., & Palevitz, B. (1999). γ‐tubulin in Chlamydomonas: Characterization of the gene and localization of the gene product in cells. Cell motility and the cytoskeleton, 42(4), 285-297.
Smith, E. F. (2002). Regulation of flagellar dynein by calcium and a role for an axonemal calmodulin and calmodulin-dependent kinase. Mol Biol Cell, 13(9), 3303-3313. https://doi.org/10.1091/mbc.e02-04-0185
Stephens, R. E., & Edds, K. T. (1976). Microtubules: structure, chemistry, and function. Physiol Rev, 56(4), 709-777. https://doi.org/10.1152/physrev.1976.56.4.709
Tam, L. W., Dentler, W. L., & Lefebvre, P. A. (2003). Defective flagellar assembly and length regulation in LF3 null mutants in Chlamydomonas. J Cell Biol, 163(3), 597-607. https://doi.org/10.1083/jcb.200307143
Upadhyaya, S., & Rao, B. J. (2019). Reciprocal regulation of photosynthesis and mitochondrial respiration by TOR kinase in Chlamydomonas reinhardtii. Plant Direct, 3(11), e00184. https://doi.org/https://doi.org/10.1002/pld3.184
Voloshin, R. A., Rodionova, M. V., Zharmukhamedov, S. K., Nejat Veziroglu, T., & Allakhverdiev, S. I. (2016). Review: Biofuel production from plant and algal biomass. International Journal of Hydrogen Energy, 41(39), 17257-17273. https://doi.org/https://doi.org/10.1016/j.ijhydene.2016.07.084
Wei, Q., Ling, K., & Hu, J. (2015). The essential roles of transition fibers in the context of cilia. Curr Opin Cell Biol, 35, 98-105. https://doi.org/10.1016/j.ceb.2015.04.015
Wheeler, G. L. (2017). Calcium-Dependent Signalling Processes in Chlamydomonas. In M. Hippler (Ed.), Chlamydomonas: Molecular Genetics and Physiology (pp. 233-255). Springer International Publishing. https://doi.org/10.1007/978-3-319-66365-4_8
Yanagisawa, H. A., Mathis, G., Oda, T., Hirono, M., Richey, E. A., Ishikawa, H., Marshall, W. F., Kikkawa, M., & Qin, H. (2014). FAP20 is an inner junction protein of doublet microtubules essential for both the planar asymmetrical waveform and stability of flagella in Chlamydomonas. Mol Biol Cell, 25(9), 1472-1483. https://doi.org/10.1091/mbc.E13-08-0464
Yang, P., Diener, D. R., Yang, C., Kohno, T., Pazour, G. J., Dienes, J. M., Agrin, N. S., King, S. M., Sale, W. S., & Kamiya, R. (2006). Radial spoke proteins of Chlamydomonas flagella. Journal of cell science, 119(6), 1165-1174.
Yokoyama, R., O’Toole, E., Ghosh, S., & Mitchell, D. R. (2004). Regulation of flagellar dynein activity by a central pair kinesin. Proceedings of the National Academy of Sciences, 101(50), 17398-17403. https://doi.org/doi:10.1073/pnas.0406817101
Zou, Y., Zeng, Q., Li, H., Liu, H., & Lu, Q. (2021). Emerging technologies of algae-based wastewater remediation for bio-fertilizer production: a promising pathway to sustainable agriculture. Journal of Chemical Technology & Biotechnology, 96(3), 551-563. https://doi.org/https://doi.org/10.1002/jctb.6602