Chemical Analysis of Amoebae
Hailey Jukes, Adele Omichinski, Nicholas Da-Costa-Bastidas, Gil Qin
Abstract
Amoebae wield a fascinating array of chemical tools to increase their chances of survival and optimize their interactions with their environment. A selection of these tools will be presented and explained in this article, so that as many people as possible may gain inspiration from the ingenuity of nature’s design solutions found in amoebae. The chemical composition of the amoeba, from its cell surface to its test, yields information pertaining to the organism’s interactions with its environment. Examination of different components of the amoeba, such as its contractile vacuole and several of its receptor proteins, clarifies the processes underlying how the amoeba organizes its nutrition and development throughout its lifetime. The contractile vacuole also serves the function of regulating the internal state of the cell, and this is a topic that is further elaborated on when discussing how the amoeba manipulates its internal state to fight off bacterial infections and optimize its predatory practices.
Introduction
Amoebae are eukaryotic single-celled organisms that display remarkable diversity. Amoebae are not confined to a single taxonomic group, but instead span across different taxonomic categories including protozoa, fungi, and algae. These organisms have also been found to live in a wide range of aquatic and terrestrial environments, from freshwater ponds to rivers and soil. Nevertheless, they all have the ability to dynamically alter their shape through the extension and retraction of pseudopods. This unique morphological feature not only allows them to move in various environments via a process termed amoeboid movement, but it also enables them to feed via phagocytosis. In fact, most amoebae can be considered as professional phagocytes, which have the capacity to efficiently tract down and engulf bacteria as well as other microorganisms (Shi et al., 2021). From a biochemical point of view, amoebae have emerged as a model organism for investigating a number of phenomena in cell biology. Their simplicity in terms of cultivation and manipulation, along with their large and easily observable single-celled structure, make them excellent experimental subjects. Researchers have used amoebae to investigate cellular functions such as phagocytosis, motility, chemotaxis and signal transduction (Lorch, 1973). Despite only being unicellular organisms, amoebae carry out numerous chemical processes as part of their life cycle. This paper will give of an overview of a subset of these processes. We will start by presenting the chemical makeup of the amoeboid cell surface and how this specific composition contributes to the organism’s function and survival. For example, this includes the surprising ability of shelled (or testate) amoebae to resist to wildfires. Continuing with the importance of the chemistry of the cell surface composition, we will discuss how specific proteins integrated in the plasma membrane allow amoebae to sense chemical gradients in their environments and initiate directed movement by a process called chemotaxis. We will also introduce how these proteins play a role in predation and phagocytosis. We will look at their iron metabolism defense mechanism against pathogenic bacteria. Taken together, these discussions will give insight into the complexity of the ongoing chemical processes within amoebae.
The importance of chemical composition in the function and survival of amoebae
Specific chemical design solutions serve as unique evolutionary adaptations for amoebae and allow them to adapt to and thrive in their environments. Examples of these adaptations include the chemical makeup of the amoeba cell surface, the formation of resistant amoeboid tests (shells) for wildfire survival, and the chemical composition of specific organelles, which all contribute to critical biological processes of the organism and function to improve its efficiency.
Chemical composition of the amoeboid cell surface
Chemical analysis of the amoeboid cell surface reveals the presence of specific carbohydrates, lipids, and proteins (Korn & Wright, 1973). The surface is structurally complex, notably containing a high abundance of mannose receptors – mannose is a carbohydrate monomer and a structural isomer of glucose – and anionic phosphate groups that contribute negative charges. This composition allows the amoebae to adsorb a wide variety of cations through phagocytic and pinocytotic events, because the cations readily bind to the anionic phosphate groups due to electrostatic attraction.
In addition, the intracellular presence of cations such as Mg2+ and Ca2+ potentiates the amoeba by creating an electric potential or voltage difference across the cell membrane, and such potentiation of amoebae has demonstrated to enhance the predatory capacity of amoeba (Fu & Casadevall, 2018) (Figure 1).
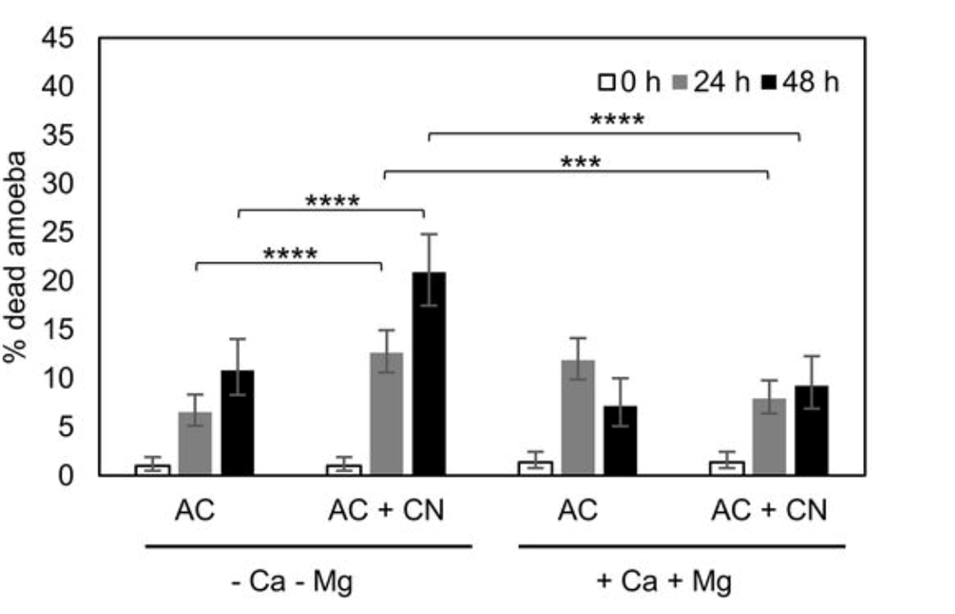
Figure 1: Plot comparing the survival of amoeba A. castellanii and C. neoformans in the presence and absence of Ca2+ and Mg2+. Calcium and magnesium cations enhance the predatory behavior of amoeba and thereby increase their rate of survival, quantitatively illustrated as a lower percentage of dead amoeba (Fu & Casadevall, 2018).
Both cations are important for the facilitation of cellular adhesions, organismal mobility, and phagocytosis. These features are linked to the predatory abilities of the amoeba and thus explain the increase seen in Figure 1 (Fu & Casadevall, 2018). The presence of the divalent cations also correlates to an increase in the surface area of the amoebae cells, allowing more efficient nutrient absorption and gas exchange, though the exact mechanism of action of this experimentally observed phenomenon has not yet been deduced.
The mannose receptors, or amoeboid mannose binding proteins, on the cell surface, recognize mannose in the organism’s surroundings and are an important component in the food gathering of amoebae. It is demonstrated that amoeba preferentially initiate endocytosis when they encounter mannose molecules in their environments, as shown in Figure 2 (Allen & Dawidowicz, 1990). The specificity of this carbohydrate-receptor interaction allows the organism to selectively endocytose its prey based on their chemical contents.
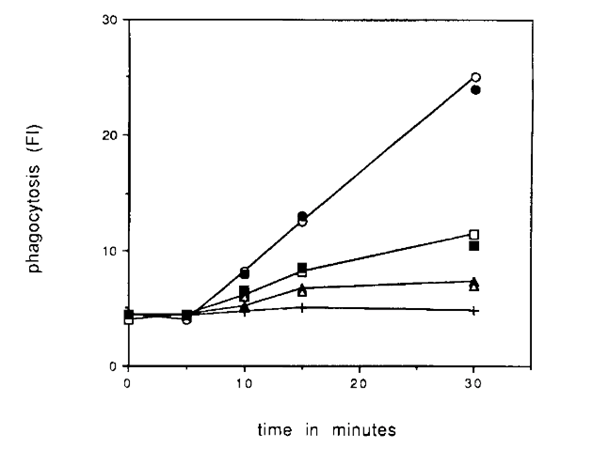
Figure 2: Graph demonstrating phagocytotic events vs. time for an amoeba in the presence of different sugars: cell cultures containing mannose is depicted as the top (steepest) trendline, which illustrates an increase in phagocytotic events in the presence of higher concentrations of mannose, thereby demonstrating the role of mannose receptors on amoeba cell surface in nutrient intake (Allen & Dawidowicz, 1990).
While they are essential in promoting amoeboid cellular nutrition events, mannose receptors also allow for the recognition of mannosylated cell walls, a key characteristic of several distinct fungal species (Ferreira et al., 2022) (Allen & Dawidowicz, 1990). Some species of amoebae possess a symbiotic relationship with several fungal pathogens. These amoebae act as natural reservoirs for the fungi, while the fungi have evolved methods to avoid digestion by the amoeba. When the amoebae infect larger mammals, the fungi can be released into the mammalian host to begin their infection of the host. This allows both pathogens to increase their probability of survival in the given host. Consequently, the mannose-binding protein acts as a notable contributor to the pathogenic mechanisms of the amoeba through the creation of a symbiotic relationship with fungal species.
Amoeboid test composition for wildfire resistance
Testate amoebae are generally known to populate peatlands, which are subject to intermittent burning by wildfires (Qin et al., 2017). This is especially relevant as recent climate change studies show an increase in the occurrence of wildfires. For amoeba that develop outer shells (referred to as tests) or testate amoebae, it is deduced that habitat burning leads to a shift in the testate amoeba population to a higher proportion of amoebae with tests constructed of xenosomes, as opposed to an equal distribution of amoebae with idiosomic and xenosomic tests (Marcisz et al., 2019). Idiosomic refers to tests consisting of organismally-secreted particles, such as biosilica or calcium carbonate. Xenosomic tests are those consisting of recycled mineral or organic particles found in the surrounding environment (Armynot du Châtelet et al., 2015). It is hypothesized that the shift in test composition of the population is due to the greater vulnerability of idiosomic tests to extreme heat (Marcisz et al., 2019). The xenosomic tests are better suited to survive wildfire conditions and the corresponding taxa of testate amoeba have adapted this design solution to optimize their chances of survival (Figure 3).
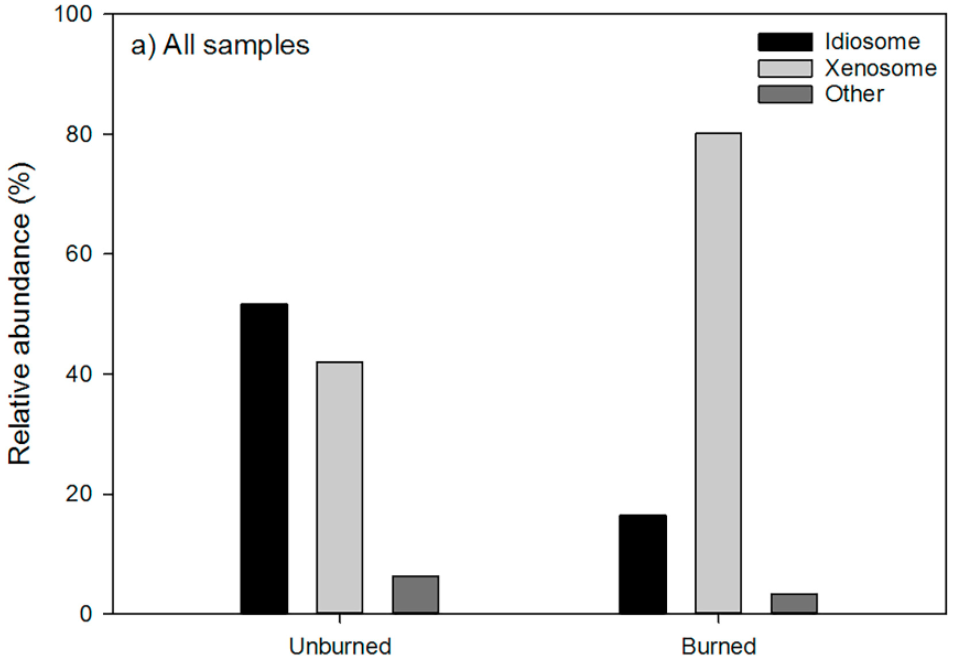
Figure 3: Comparison of population abundance of idiosomic and xenosomic testate amoeba before and after habitat burning. It is demonstrated that after an event of habitat burning, the relative abundance of amoeba with xenosomic tests increased, while the relative abundance of amoeba with idiosomic tests decreased. This is evidence of xenosomic tests having a higher resistance to heat than idiosomic tests (Qin et al., 2017).
Studies show that xenosomic tests are thicker and more robust than ones containing idiosomes in areas where there are sufficient minerals available (Armynot du Châtelet et al., 2015). This is because these amoebae have more freedom to select particles of different shapes and sizes to form their three-dimensional shells (Figure 3). The contents of these xenosomic shells also tend to have better decay resistance due to their composition.
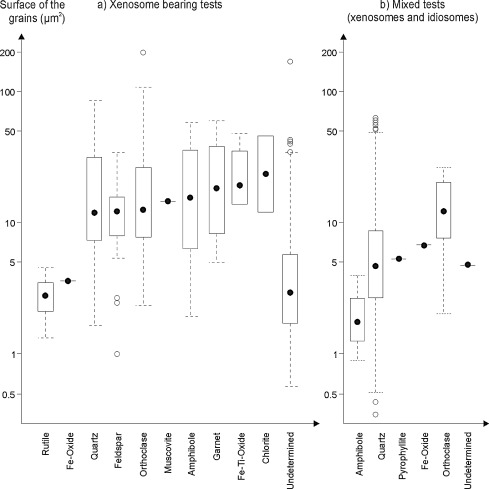
Figure 4: Graphical comparison of test area for xenosomic tests vs. idiosomic and xenosomic tests shows that, on average, the test thickness/grain size of xenosomic tests is larger but is dependent on the minerals available (Armynot du Châtelet et al., 2015).
In peatlands there are a wide variety of minerals present, therefore the xenosomic test formation technique is well suited (Qin et al., 2017). As the grains available to the xenosomic testate amoeba are environmentally specific, the xenosomic tests are more likely to survive wildfires and extreme heat events; they consist of minerals that have likely have resistance to these conditions. Therefore, the amoeboid shells are specifically chemically designed for the survival and proliferation of the organism.
The ameboid contractile vacuole
In amoebae, the contractile vacuole (which is specific to the organism) is specialized, allowing them to thrive in their habitats. The contractile vacuole functions as the excretory, or waste removing site of the organism, and most notably, it removes excess water from the cell to allow osmoregulation (Bairati & Lehmann, 1953).
The function of the contractile vacuole is directly related to its chemical composition. The contractile vacuole is used to segregate hypotonic solutions from the rest of the cytoplasm and discharge them outside of the cell to maintain internal homeostasis (Du et al., 2008). This function is more often performed when the organism is in a hypotonic environment, which is common for lake or freshwater-dwelling amoebae. Hypotonic environments cause water to flow into the (relatively) hypertonic amoeboid cells, lowering their solute concentration. This then affects the internal pH of the organism, which impacts the metabolic reactions that require a specific pH and can denature proteins, since changes in pH may alter the charge of molecules. The mechanism of the contractile vacuole relies on active transport of cytoplasmic ions into the vacuole to create a concentration gradient (Komsic-Buchmann et al., 2014) (Bairati & Lehmann, 1953). This concentration gradient causes water to flow from the cytoplasm into the vacuole to equilibrate solute concentrations, and thus water is removed from the cytoplasm and the internal concentration of ions is maintained. The contractile vacuolar membrane contains a high number of protein channels known as aquaporin major intrinsic protein 1 (MIP1). This protein ensures that only a very small osmotic gradient is required to cause a water flux into the vacuole, thus homeostasis can be closely regulated (Benga, 2012). It is highly selective for only water molecules, due to the specific shape and charge states of the pore (Figure 5). The pore bends at a 25-degree angle and varies in its diameter to exclude larger and smaller molecules. The entrance to the aqueous pathway is composed of charged and polar residues to attract water molecules, whereas the interior is non-polar. The feature allows for the uninterrupted flow of water through the pore, causing the transport to be faster (Benga, 2012). Additionally, light microscopic immunofluorescence has confirmed the presence of calmodulin (a calcium-ion binding protein that acts a signaling protein) in the vacuolar membrane, along with a high density of proton pumps (Heuser et al., 1993) (Komsic-Buchmann et al., 2014). These components allow the cell to harness the potential energy from a proton motive force to aid in moving water into the contractile vacuole.
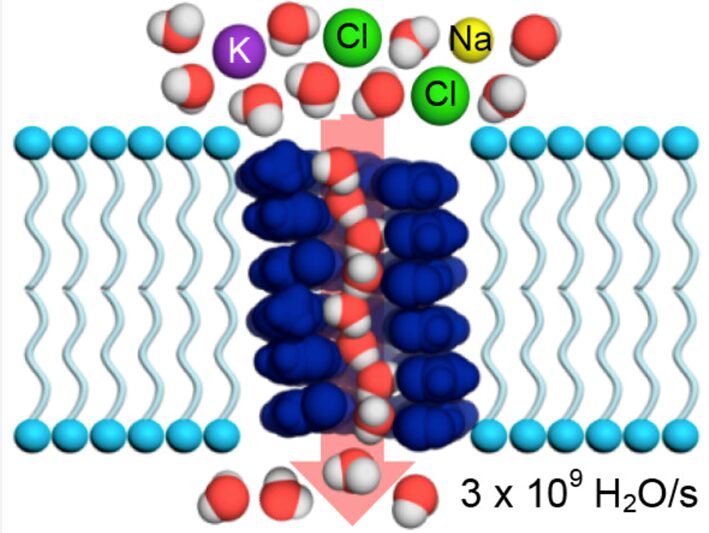
Figure 5: Model of the Aquaporin 1 aqueous pathway present in the contractile vacuolar membrane of amoebae. The pore has a 25-degree angle bend and varies in size to select for water molecules and exclude all others. The interior of the channel is non-polar, allowing the uninterrupted flow of water (Shen et al., 2020).
When the contractile vacuolar membrane reaches a threshold (maximum) diameter, cellular proteins called LvsA and Dranin are recruited to the vacuolar membrane (Heuser et al., 1993). These proteins facilitate the fusion of the vacuolar membrane with the cell membrane, and the release of its contents into the extracellular space. Studies suggest that the composition of the vacuole, and its distinct membrane components, allow for it to collapse and flatten against the plasma membrane to release its contents without completely fusing. This allows for the recycling of the vacuole and saves the organism the energy and effort required to synthesize another. This shows that the design and components of the contractile vacuole are highly purposeful in promoting the osmoregulation of the amoeba.
The role of G-protein coupled receptors in chemotaxing amoebae
Chemotaxis, the directed motion of cells in response to an extracellular chemical gradient, is a fundamental biological process seen in many cellular organisms. It enables them to monitor the availability of life-sustaining nutrients in their extracellular environment and can also serve as a mode of communication between neighboring cells (Devreotes & Zigmond, 1988). Amoeboid cells, specifically Dictyostelium discoideum cells, provide an excellent model for studying chemotaxis since they exhibit strong and highly specific chemotactic responses. This process relies on chemoattractants binding to G-protein coupled receptors (GPCRs) on the cell’s surface, which activates downstream signaling pathways. These chemical cascades lead to the rearrangement of cytoskeletal proteins such as actin and myosin that induce the extension of pseudopods towards higher concentrations of a chemoattractant (Wu, 2005).
Overview of G-protein coupled receptor structure and function
G-protein coupled receptors make up the largest and the most diverse class of cell surface receptors in eukaryotic organisms. These integral membrane proteins mediate cellular responses to a wide range of external stimuli such as photons, ions, proteins, lipids, nucleotides, and hormones. The common structural feature of all GPCRs is the transmembrane domain, which is composed of seven alpha-helical stretches of 25 to 35 amino-acid residues that span the plasma membrane. The amino acid residues within these helical stretches are relatively non-polar, making these segments highly hydrophobic and therefore facilitating their integration into the lipid bilayer of the cell membrane. In contrast, the intracellular and extracellular loops (ICL and ECL in Figure 6) connecting the seven helices are more hydrophilic in nature. The transmembrane domain serves as a conduit for relaying signals from the extracellular amino-terminal region (ligand-binding domain of the GPCR) to the intracellular carboxy-terminal (C-terminal) region of the protein. In the absence of ligand-binding, the intracellular C-terminal region is typically bound to a heterotrimeric G-protein but may also be linked to other signaling proteins such as protein kinases and arrestins (Schiöth & Fredriksson, 2005).
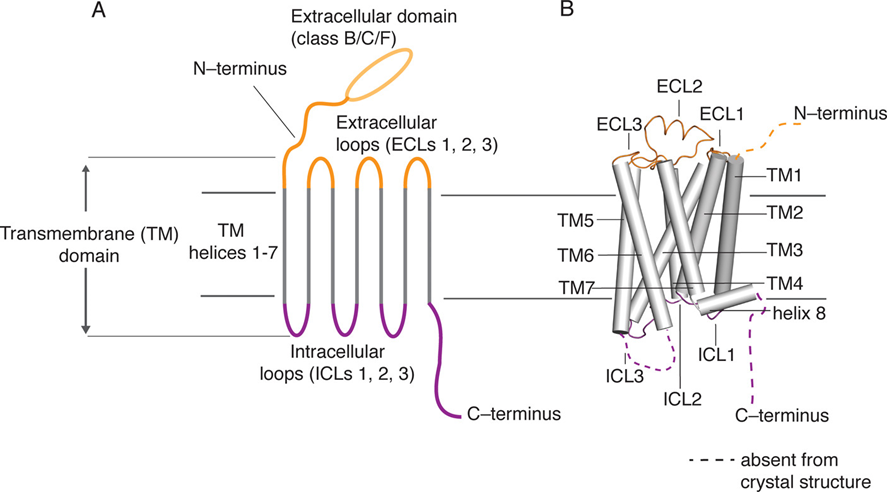
Figure 6: Schematic representation of the basic structure (A) and topology (B) of GPCRs. The seven transmembrane (TM) helices are shown in grey. The three extracellular loops (ECL) and the extracellular domain including the protein’s N-terminus are shown in orange. The three intracellular loops (ICL) and the intracellular domain including the protein’s C-terminus are shown in purple (Latorraca, Venkatakrishnan, & Dror, 2017).
The ability of a GPCR to convey a signal from the extracellular space to the inside of the cell relies on its capacity to undergo structural changes. When an external signaling molecule (ligand) binds to the extracellular domain of the GPCR, it induces a conformational change in the protein structure that facilitates the dissociation of a heterotrimeric G protein (or other signaling proteins) from the intracellular C-terminal domain of the GPCR. The G-protein can then be involved in regulating signal transduction in response to stimulus (Latorraca, Venkatakrishnan, & Dror, 2017)
Heterotrimeric guanine nucleotide-binding regulatory proteins (G proteins) are important molecular switches in the regulation of GPCR signaling. They each consist of three different subunits: alpha, beta and gamma (Syrovatkina et al., 2016). The alpha subunit can bind to either guanosine triphosphate (GTP) or guanosine diphosphate (GDP) depending on whether the G protein is in an active state (GTP-bound) or an inactive state (GDP-bound). In the absence of an external signal, the GDP-bound Ga subunit forms a complex with the Gb and Gg subunits, which is attached to a GPCR. Upon ligand binding, the conformational changes in the GPCR structure promote the activation of the heterotrimeric G-protein by exchange of GDP for GTP on the alpha subunit. This causes the dissociation of the G-protein complex into a GTP-bound Ga subunit and a Gbg heterodimer (Baltoumas, Theodoropoulou, & Hamodrakas, 2013). Both components remain anchored to the cell membrane (the Gbg dimer is attached via the Gg subunit), but dissociate from the GPCR (Figure 7), allowing them to activate other small G proteins that also switch from an inactive GDP-bound state to an active GTP-bound state. These smaller proteins can only interact with downstream effectors in the active GTP-bound state (Kamato et al., 2015).
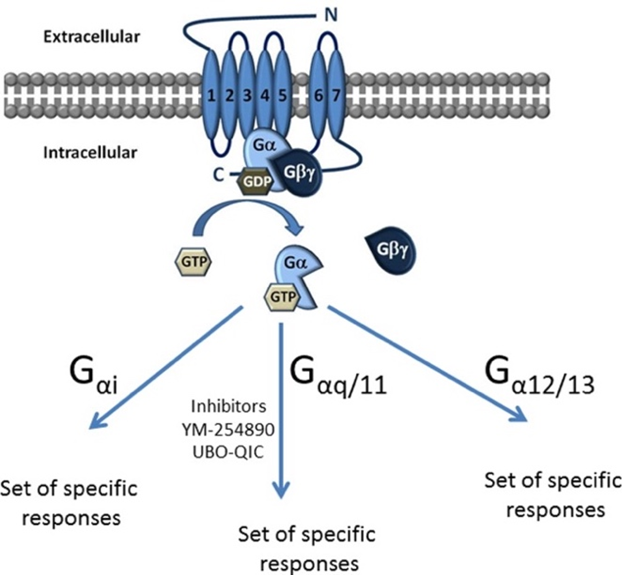
Figure 7: Schematic representation of the inactive GDP bound state and the active GTP bound state of the GPCR. According to the sequence and functional similarities of their Ga subunit, G proteins can be classified into four different families: Gas, Gai, Gaq and Ga12/13. The Gas and GaI families are involved in the regulation of adenyl cyclase activity, whereas Gaq activates phospholipase Cb and Ga12/13 is responsible for stimulating small GTPase activity (Kamato et al., 2015).
Chemotactic responses in Dyctyostelium discoideum
Dictyostelium discoideum demonstrates a remarkable duality in their chemotactic responses. When preferred food sources, such as bacteria, are abundant in their environment, these cells employ specific GPCR’s to control directed movement towards folic acid secreted by bacteria. These receptors not only help the cells locate and chase prey but are also essential for the engulfment of bacteria through phagocytosis. Conversely, in the condition of nutrient scarcity and starvation, Dictyostelium shift their behavior, transitioning from a predatory mode to a cooperative mode. These amoeboid cells rely on GPCR’s specific to cyclic adenosine monophosphate (cAMP) to orchestrate the aggregation of cells into colonies that ensure the species’ long-term survival (Figure 8). This duality highlights the ability of Dictyostelium to adapt its chemotactic responses based on the availability of nutrients and its life stage, transitioning from solitary predation to cooperative multicellular development (Liu et al., 2018).
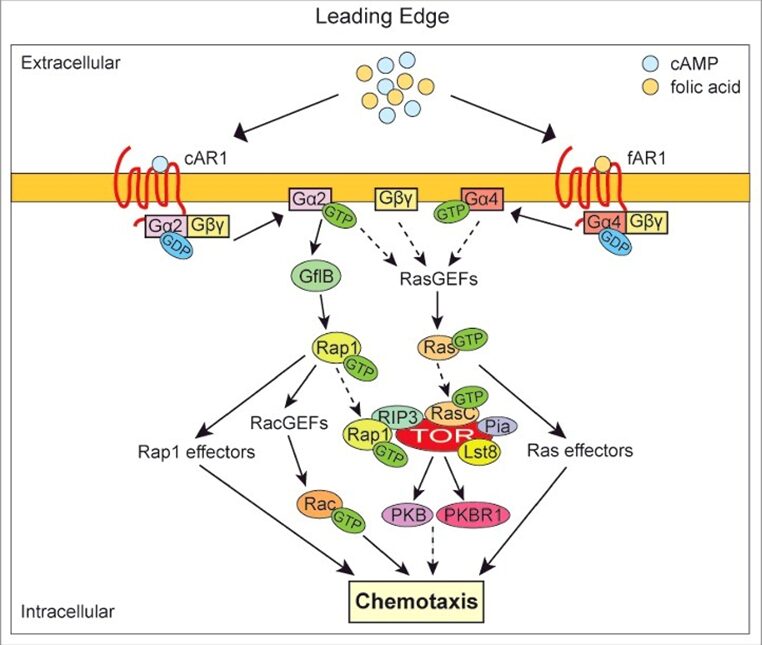
Figure 8: Diagram summarizing the GPCR-mediated pathways that regulate chemotactic responses in Dictyostelium discoideum. The left side shows the chemotaxis response initiated by the binding of cAMP to cAR1 (cAMP receptor) and the right side shows the chemotaxis response initiated by the binding of folic acid to fAR1 (folic acid receptor). In both cases, the binding of the chemoattractant to its respective GPCR leads to the dissociation of the heterotrimeric G-protein into separated Ga-GTP unit and Gbg dimer. These interact with various effectors, which activate a series of downstream signaling pathways ultimately resulting in chemotaxis (Liu et al., 2018).
Chemoattraction to cAMP in Dictyostelium discoideum
In nutrient-depleted environment, Dictyostelium discoideum cells undergo a developmental program, which leads to the aggregation of cells into colonies and the morphogenesis of multicellular fruiting bodies. This process relies on the ability of starved cells to synthesize and secrete pulses of cAMP, derived from adenosine triphosphate (ATP). These cAMP pulses establish an extracellular cAMP gradient around the secreting cells and function as a chemoattractant to guide the migration of neighboring cells. Neighboring cells detect this chemical gradient through cAMP specific GPCR receptors on their cell surface and orient their movement towards the cAMP secreting cells. The chemotaxing cells also begin to secrete their own pulses of cAMP, creating a propagating wave that repeats in approximately six-minute intervals (Consalvo et al., 2019). This coordinated movement leads the cells toward a central aggregation site, where they form a multicellular mound, known as a “slug” comprising anywhere from 10 to 105 cells. This slug will crawl towards light and warmth before forming into a fruiting body. Ultimately, the fruiting body will differentiate into spores, stalk cells and supporting structures that support the stalk and spore head. Once the spores are reintegrated into a nutrient-rich habitat, they will germinate and resume proliferation as individual amoebae, concluding the amoeba’s life cycle (Figure 9) (Kim, Haastert, & Devreotes, 1996).
The Dictyostelid group of social amoebas, which includes D. discoideum, developed this multicellularity 600 million years ago as a survival tactic in response to starvation. Their ability to form spores and stalk cells with specialized roles like basal disk cells, upper cup cells, and lower cup cells, demonstrates a high level of organization in their multicellular development. Furthermore, multicellularity by the morphogenesis of a fruiting body has evolved independently multiples times across various eukaryotic and prokaryotic lineages. This suggests its significance as a successful strategy for survival in nutrient depleted environments. Specifically, Dictyostelid amoeba seem to have achieved a particularly advanced level of organization in this process, characterized by efficient cellular aggregation and highly regulated morphogenesis facilitated by cAMP signaling (Schaap, 2016).
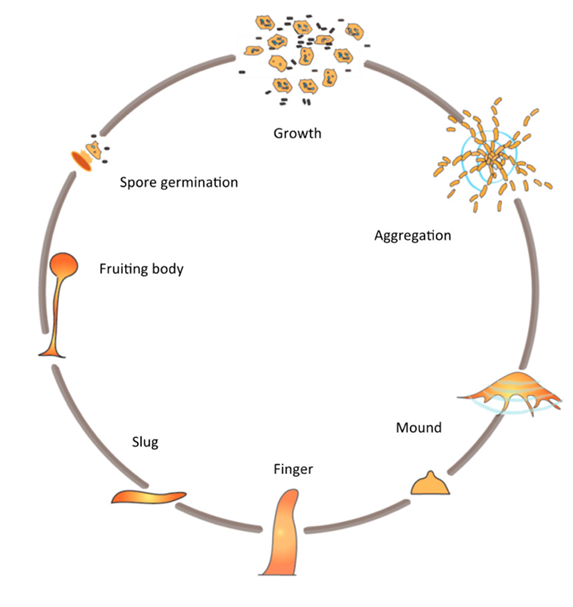
Figure 9: Scheme of the life cycle of Dictyostelium discoideum cells. D. discoideum, as individual cells, aggregate towards each other to form a cell cluster, which undergoes various developmental phases, ultimately leading to the emergence of a fruiting body housing spores (Domínguez-Martín et al., 2017).
Specific G-proteins coupled receptors involved in cAMP detection
In Dictyostelium, extracellular cAMP is detected by cell surface GPCR’s. These amoeboid cells express four distinct cAMP receptors, cAR1, cAR2, cAR3 and cAR4, each of which are expressed at different stages of cellular development (Figure 10). Of these, cAR1 stands out as the primary receptor involved in cAMP chemotaxis and was actually the first chemoattractant receptor to be discovered in eukaryotic cells (Figure 11). cAR1 is maximally expressed while cells are aggregating, whereas the three other cAMP receptors, are all expressed at later developmental stages. cAR3 expression levels rise towards the end of cell aggregation, whereas cAR2 and cAR4 expression are maximal in the slug and fruiting body stages. However, it has been shown that there is a partial redundancy between cAr1 and cAR3, as both are expressed during early developmental stages. In fact, cells tested with a gene deletion of cAR1 were surprisingly still able to respond chemotactically to cAMP, but with diminished sensitivity, and this was due to the activity of cAR3. In addition, cells having both cAR1 and cAR3 deleted lacked the ability to aggregate in response to cAMP (Hereld & Devreotes, 2004).
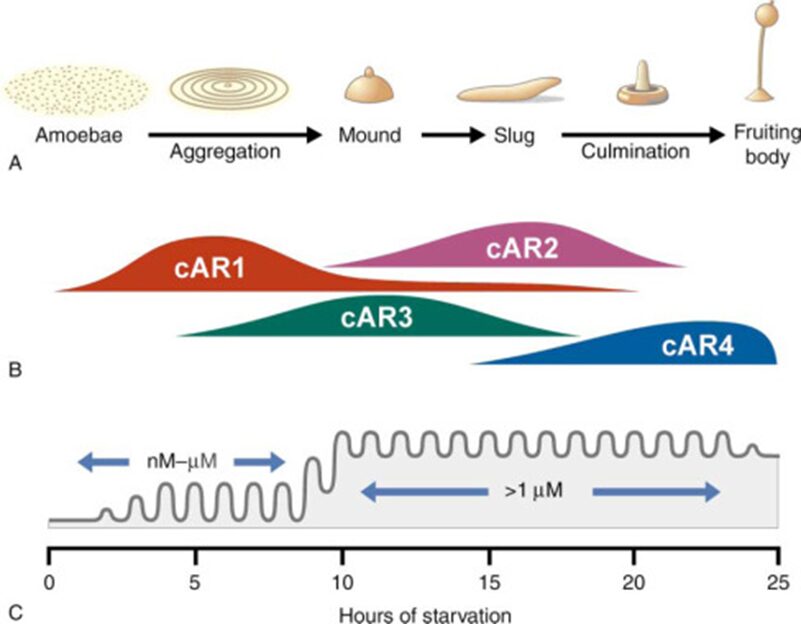
Figure 10: Scheme of the 24 hour developmental program of Dictyostelium discoideum demonstrating the correlation between (A) the developmental stage morphology (B) cAR expression and (C) concentration level of extracellular cAMP (Hereld & Devreotes, 2004).
Despite contributing at different stages of the life cycle, all four cAMP receptors are linked to the same signal-transduction pathways inside the cell, which mediates activation of adenylyl cyclase (converts ATP into cyclic AMP) and actin polymerization. The varying affinity of the four receptors for cAMP enables them to intervene appropriately in specific environmental conditions (varying cAMP levels). As cells draw close, they sequentially express cAR receptors with lower affinity, achieving the appropriate response by switching receptor subtypes while preserving similar mechanisms of signal transduction (Kim, Haastert, & Devreotes, 1996).
Sustained activation of cAR1 triggers a series of events resulting not only in excitation but also desensitization of the response to cAMP. The activated receptors undergo desensitization by adaptation to the cAMP gradient and by loss of ligand binding. In order to prevent the extracellular cAMP concentration from accumulating and saturating the cAMP sensing mechanism, aggregating cells additionally secrete cAMP-degrading phosphodiesterases. These enzymes play a crucial role in maintaining the appropriate cAMP concentration and the loss of phosphodiesterases can even inhibit aggregation of Dictyostelium cells (Consalvo et al., 2019). In addition, aggregation of Dicytostelium cells can be inhibited by the disruption of Ga2, the Ga subunit that interacts with the cAMP receptor cAR1. Without proper functioning of Ga2, cells do not respond to the chemoattractant cAMP and are therefore unable to aggregate (Liu et al., 2018).
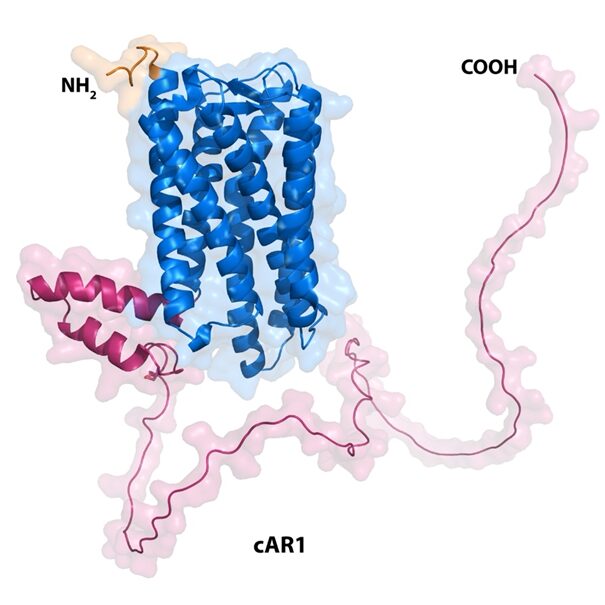
Figure 11: Structure of the cAMP GPCR involved in chemotaxis in amoebae. Surface representation of the AlphaFold model of the cAR1 receptor (PDB code number AF-P13773-F1-model_v4) from D. discoideum highlighting the extracellular ligand binding region (orange), the 7-transmembrane helical region (blue) and the intracellular G protein binding region (magenta). The figure was made using the PyMOL software (Jumper et al., 2021).
Chemoattraction to folic acid
As previously touched upon, cAMP is one of the two main chemoattractants Dictyostelium discoideum responds to. Indeed, this organism also engages in chemotaxis along the concentration gradient of folic acid. While cAMP-caused chemotaxis is linked to the development of the amoeba and its activity at different stages of its life cycle, folic acid-caused chemotaxis is instead linked to predation and thus nutrition for the amoeba.
Folate (a name used interchangeably with folic acid) is a bacterial metabolite, and thus its detection in the environment signals the presence of bacterial prey for D. discoideum, naturally causing the amoeba to follow the concentration gradient to find its food source (Pan et al., 2018).
Similar to cAMP chemotaxis, folate chemotaxis is mediated by GPCRs in the cell membrane. Although scientists have known that D. discoideum moves along folate concentration gradients for decades, it was only in 2016 that specific receptors for this molecule were identified (Pan et al., 2016). Two were discovered, and researchers labelled them fAR1 and fAR2. It is now known that fAR1 is the most important of the two receptors, as research conducted on far2-null strains of D. discoideum, meaning strains of the organism that have no active fAR2 receptors, showed that the amoebae were still able to follow folate concentration gradients and bind to bacteria to engage in phagocytosis (Pan et al., 2016). On the other hand, far1-null strains were unable to respond to folate in their environment, thus establishing the importance of this receptor in amoeboid chemotaxis to folate (Pan et al., 2016).
A more detailed examination of the signaling pathways leading to chemotaxis
As fAR1 is a GPCR, it has the same general structure as presented above in relation to cAMP chemotaxis, possessing seven transmembrane domains, one amino-terminal extracellular domain, and one carboxyl-terminal intracellular domain (see Figure 12 below) (Pan et al., 2018). This can be seen below in Figure 12. Research into the extracellular domain has shown that it folds into a Venus Fly Trap (VFT) structure, and it is this structure that folic acid binds to during activation of the signaling pathways leading to chemotaxis (Pan et al., 2018).
As previously mentioned, the binding of the folate ligand to the GPCR leads to a series of conformational changes that result in the phosphorylation of the heterotrimeric G-protein bound to the receptor on the inside of the cell. The signaling pathways refer to the subsequent rapid activation of small G-proteins within the cell, which will go on to cause downstream signaling events. Since small G-proteins are more strongly activated at the leading edge of the chemotactic cell than at other parts of the cell, an intracellular G-protein activation gradient is created (Liu et al., 2018).
Although the activation of the heterotrimeric G-proteins bound to the receptors is proportional to the steepness of the extracellular chemical gradient, it has been observed that the activation of small G-proteins at the leading edge of the cell is much stronger than the steepness of that same gradient, thus leading to an even steeper intracellular small G-protein gradient (Liu et al., 2018). In other words, the extracellular signal is amplified along the signaling pathway within the cell.
The intracellular gradient causes changes in the structure of the cytoskeleton which explain the phenomenon of chemotaxis. Indeed, actin polymerization is promoted at the leading edge of the cell, and so is the formation acto-myosin filaments at the rear of cell. This in turn leads to the formation of new pseudopods in the direction of movement, and the inhibition of their formation in the opposite direction (Liu et al., 2018).
This process is common to chemotaxis in response to both cAMP and to folic acid. However, it is important to note that the Ga subunits of the G-proteins linked to the GPCRs mediating each type of chemotaxis are different. This allows for different responses to different chemoattractants, even though they both utilize G-proteins, employ very similar signaling pathways, and have the same Gbg subunits.
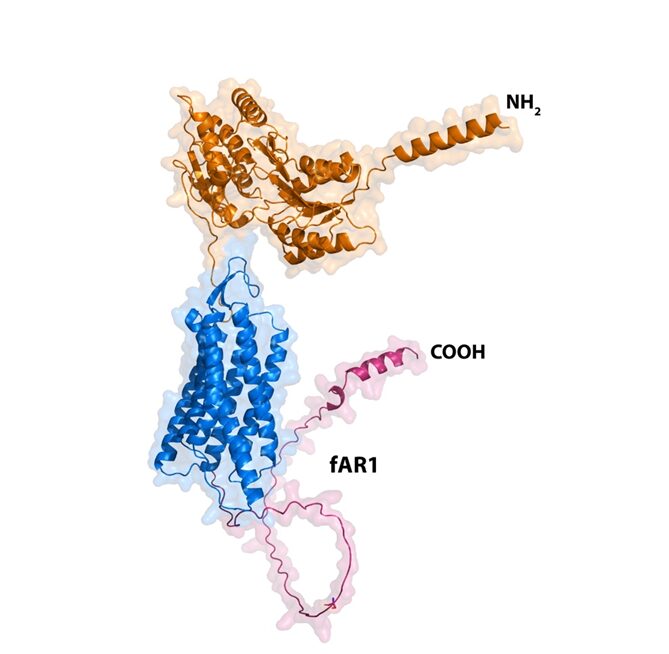
Figure 12: Structure of the folic acid GPCR involved in chemotaxis in amoebae. Surface representation of the AlphaFold model of the fR1 receptor (right; AF-Q54U89-F1-model_v4) from D. discoideum highlighting the extracellular ligand binding region (orange), the 7-transmembrane helical region (blue) and the intracellular G protein binding region (magenta). The figure was made using the PyMOL software (Jumper et al., 2021).
GPCRs and phagocytosis
The process of predation by phagocytic cells is commonly broken down into two main steps. First, the cells make use of certain receptors, commonly believed to be GPCRs, to detect a chemical gradient signaling the presence of prey, and they follow this gradient until the prey is found. Then, once they come into contact with the prey, the cells use a second receptor to recognize and engulf it via phagocytosis. These second receptors are broadly labelled pattern recognition receptors (PRRs), as their role is to detect a common pattern to the phagocyte’s prey on the surface of the cells (Xu et al., 2021). In the case of bacteria, these patterns are often called microbial-associated molecular patterns (MAMPs) (Pan et al., 2018).
However, even after extensive research, no such PRRs were found on the surface of D. discoideum. Yet these cells are “professional phagocytes,” displaying astounding abilities to hunt down and consume bacteria. How could this be? Eventually, scientists realized that these amoebae used the exact same GPCRs involved in folic acid chemotaxis to recognize MAMPs and engage in phagocytosis (Pan et al., 2016).
The MAMP in this case is lipopolysaccharide (LPS), a static component of the bacterial coat. Indeed, the VFT domains discussed above in relation to chemotaxis also display the ability to bind to particular saccharides in the core region of LPS (Pan et al., 2018). The presence of this MAMP is essential for successful phagocytosis. Although D. discoideum cells display the beginnings of the formation of a phagocytic cup when encountering many different objects, the presence of LPS is vital for stabilizing the cup. Without it, the cup collapses before an object is halfway engulfed, as seen in Figure 13A and 13B. In contrast, objects coated with LPS are engulfed effectively and swiftly (Row C below) (Pan et al., 2018). Furthermore, as the concentration of the MAMP on the surface of the target increases, the proportion of targets phagocytosed also increases, as seen in Figure 13D.
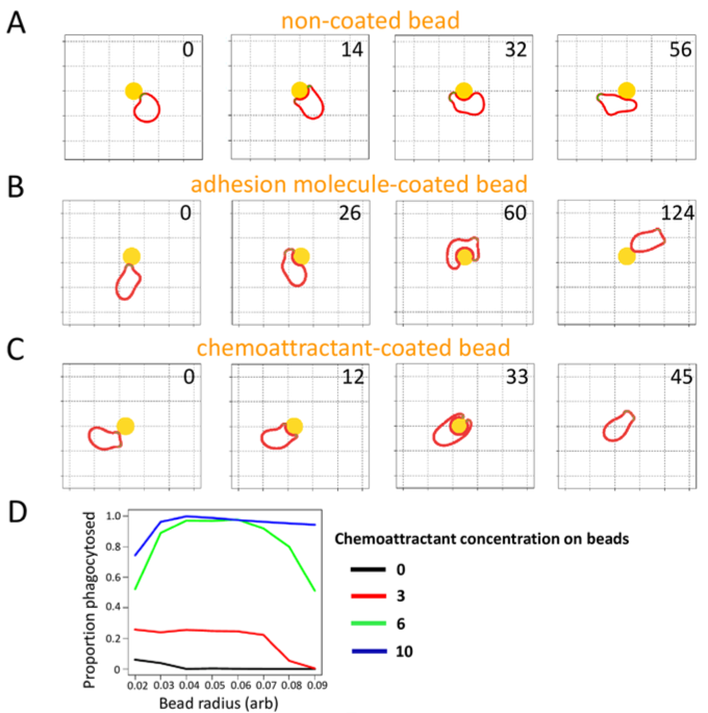
Figure 13: Comparison of phagocytic responses to different levels of chemoattractant concentration on coated beads. (A, B, C): Response of phagocyte to beads with different coatings. Each coating is shown with snapshots at different times, indicated by the number in the upper right of each snapshot. (D): Proportion of beads phagocytosed as a function of bead radius and in relation to different concentrations of chemoattractant on the beads. (Pan et al., 2018)
Phagocytosis makes use of the same GPCRs as folic acid-caused chemotaxis. It follows that the signaling pathways and reorganization of the cytoskeleton necessary for this process to be completed are also very similar to those used during chemotaxis.
It is clear that D. discoideum is a master of optimization of its cellular processes. Not only does it use the same GPCR for both chemotaxis of folic acid and phagocytosis, but it also uses similar signaling pathways within the cell to respond to both chemoattractants and the MAMPs on the surface of its prey. This organism simply exploits slight changes to sub-components involved in each process to fine-tune its reaction to different stimuli. Thus, it is able to optimize its use of cellular components and resources while retaining access to a variety of functions and behaviors. In this sense, D. discoideum is a perfect example of the law of parsimony at work and is a model to follow when seeking ways to maximize efficiency in a system.
Iron metabolism and bacterial infection resistance in amoeba
Many amoebae, for example, the soil-dwelling Dictyostelium discoideum, prey on bacteria for nutrients through phagocytosis. However, despite bacteria being a source of food for amoebae and other bacterivorous protozoa, many are also pathogenic. Consequently, amoebae, such as Dictyostelium, have evolved an infection resistance mechanism to bacterial pathogens through regulation of iron homeostasis. Iron is a vital element for amoebae and concurrently a crucial nutrient in bacterial metabolism that is critical for their survival. Therefore, iron homeostasis in amoeba, is not only important in regulating the growth of amoeboid cells, but also functions to resist pathogens through a mechanism known as iron starvation (Bozzaro and Peracino, 2013). This renders amoeba an excellent microbicidal phagocyte model for studying the cellular antimicrobial functions and cell-autonomous defense mechanisms of mononuclear phagocyte system (MPS) – namely, phagocytosis and autophagy – that have evolved from the ancient predator-prey relationships between eukaryotes and prokaryotes.
Recognition of pathogen-associated molecular patterns (PAMPs) and phagocytosis initiation
In mammals, to initiate an immune response, innate immune cells recognize PAMPs released by bacterial cells through a collection of pathogen recognition receptors that comprises toll-like receptors (TLR), integrins, scavenger receptors, and C-type lectins (Freeman and Grinstein, 2014). TLR, specifically, are responsible for constantly patrolling the endocytic compartments and recognizing PAMPs within these compartments to induce cell-autonomous defense. In comparison, lectins, scavenger receptors, and integrins serve as phagocytic receptors that can bind to particles and initiate internalization and subsequent degradation. In lower eukaryotes such as Dictyostelium, the complete TIR protein is absent, however, it does produce two cytosolic proteins containing TLR domains (TlrA and TlrB) that perform the same PAMP-recognition function. Phagocytic receptors identified in the organism belong to the integrin-like Sib protein family, such as SibA and SibC, that allows binding to substrate and particles (Dunn et al., 2018).
As seen in Figure 14, it is demonstrated that phagocytosis of non-pathogenic bacteria in Dictyostelium cells does not alter the regular pathway of phagosome maturation. That is, the phagocytosis of non-pathogenic bacteria does not prevent the recruiting of important components of the phagosomal maturation pathway to the phagocytic compartment of food sources. These components commonly include metal transporters, vacuolar ATPase (V-ATPase), lysosomal enzymes, and NADPH oxidase (NOX) complex. However, pathogenic bacteria can modify the phagosomal maturation pathway such that the cell is no longer able to transform the phagocytic compartment into a microbicidal phagosome. This is achieved by preventing the binding of vesicles bound to the NOX complex and metal transporters as well as vesicles containing lysosomal enzymes and V-ATPase to the phagosome. Such mechanism behind the inhibition of normal phagosomal maturation is explained later in the article. Once able to freely proliferate in the phagocytic compartment, the pathogen can exit the phagosome to be exocytosed or penetrate the host cell through lysis or non-lytic ejection.
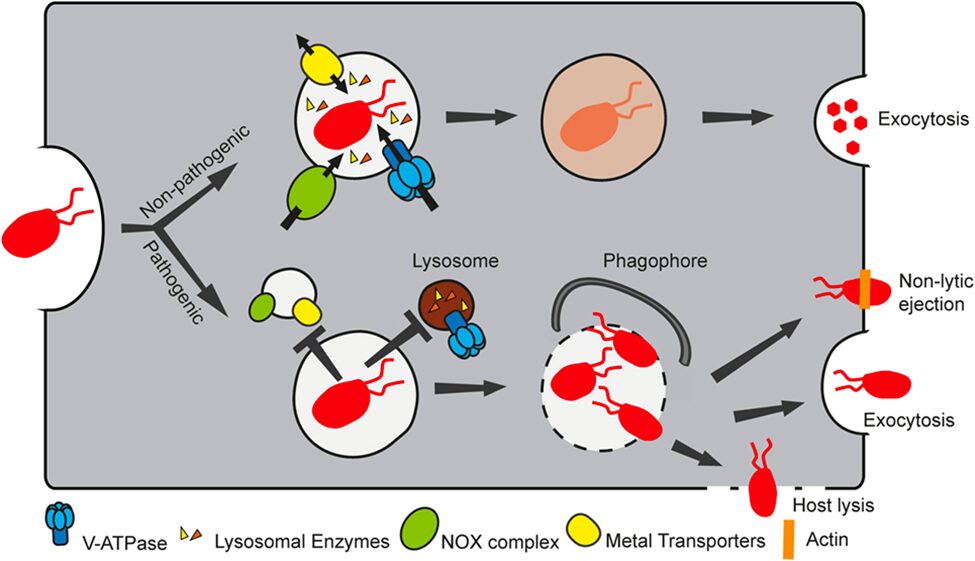
Figure 14. Non-pathogenic bacteria follow the standard phagosomal maturation pathway to be digested as a source of nutrient for the host cell. However, pathogenic bacteria change the phagosomal maturation pathway by preventing the binding of NOX complexes, metal transporters, lysosomal enzymes, and V-ATPase to the phagosome, thereby escaping the defense mechanism of the host cell (Dunn et al., 2018).
Iron metabolism in amoeba
In the iron metabolism of amoeba, the key integral membrane proteins that act as iron transporters encoded by the amoeboid genome include Nramp1 (Natural Resistance-Associated Macrophage Proteins 1) and Nramp2 (Natural Resistance-Associated Macrophage Proteins 2). Nramp1 plays an essential role in importing iron through phagocytosis and pinocytosis into the cell (Bozzaro and Peracino, 2013). In the two endocytotic processes, Nramp1 transporters are mobilized from the Golgi apparatus to the phagosomes and macropinosomes, which are endocytic structures responsible for the ingestion of extracellular fluid and the uptake of microbes, respectively. As the bacterium is degraded, iron enters the cytosol through Nramp1 from the phagosome. On the other hand, Nramp2 is a contractile vacuole membrane protein, responsible for coordinating osmosis as well as the transport of iron into and out of the vacuole. In phagocytosis and pinocytosis at the cell membrane, vacuolar (H+) – ATPase that abundantly bud the surface of contractile vacuoles are recruited with phagosomes and macropinosomes. The membrane potential generated by V-H+ ATPase is important in regulating the transport of iron by Nramp1 and Nramp2. It is theorized that Nramp iron transporters are proton-coupled symporters where the transport of ferrous ions to the cytosol are coupled with the pumping of protons into the phagocytic compartment by active transport (Forbes and Gros, 2001).
However, Nramp1 is specific in transporting only the divalent ferrous ion, while the digestion of bacteria in phagosomes results in ferric ions that require reduction before transporting into the cytosol. Indeed, ferric reductases belonging to the domon-cytB561 family are encoded in the amoeba genome to reduce ferric to ferrous ions to be ready for cotransport via Nramp1 (Bozzaro and Peracino, 2013). Inside the cell, the majority of the ferrous ions are transported via another iron transporter – mitoferrin – to the mitochondria, while excess cytosolic iron is stored in the vacuole. The iron stored in the vacuole is released through the exocytotic fusion of the vacuole with the membrane or – if the amoeba is in a starving condition where only intracellular iron is available – returns to the cytosol through Nramp2, which actively transport ferrous ions into the cytosol from the vacuole via the same proton-coupled cotransport as Nramp1.
Iron starvation and pathogen resistance
In a study done by Bozzaro and Peracino, the researchers demonstrated that a disruption to the Nramp gene, responsible for encoding Nramp1 and Nramp2 transporter proteins, results in an enhanced proliferation and growth of bacteria such as Legionella and Mycobacteria in cells of Dictyostelium, thus increasing the amoeba’s proneness to bacterial infections. In contrast, the overexpression of Nramp1 has been demonstrated to reduce intracellular growth of Legionella.
A possible mechanism of host defense deployed by the amoeba host cell is to deprive the pathogenic bacteria of iron through iron depletion in phagosomes via the efflux of iron via iron transporters such as Nramp1 (Figure 15, left). This mechanism is known as metal deprivation or nutritional immunity (Dunn et al., 2018).
In contrast, as seen in Figure 15, the pathogenic bacteria, L. pneumophila, is observed to inhibit the Nramp1 activity by inhibiting the fusion between the acidic vesicle containing V-H+ ATPase and the endocytotic structure. The inability of the cell to recruit V-H+ ATPase to the macropinosome or phagosome results in an inability of Nramp1 to pump ferrous ions out to the cytosol, since Nramp1 transporters function through proton coupled symport. In other words, by inhibiting the co-localization of Nramp1 and V-H+ ATPase in the phagosome or macropinosome, Nramp1 is prevented from facilitating proton-driven ferrous ion efflux into the cytosol. Instead, cytosolic ferrous ions will enter the phagosome or macropinosome through passive transport, providing excess nutrients for the pathogens, further facilitating their growth and proliferation.
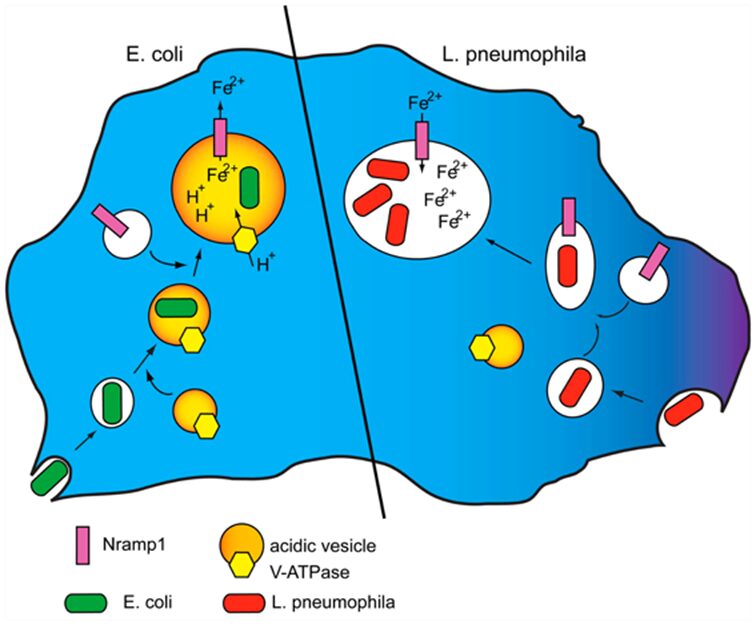
Figure 15: Left: V-H+ ATPase and Nramp1 are mobilized by phagosomes upon the intake of bacteria such as E. coli. Then, the phagosome is acidified as the V-H+ ATPase imports protons into the phagosome, resulting in a proton gradient that transports divalent iron from the phagosome into the cytosol, depriving bacteria in the phagosome of necessary iron. Right: Amoeba intakes L. pneumophila through macropinocytosis. The pathogenic mechanism of L. pneumophila involves preventing the macropinosome to fuse with the V-H+ ATPase-bearing acidic vesicle. After Nramp1 is recruited to the macropinosome, there is no proton gradient generated by active transport in the macropinosome, which means that iron efflux via Nramp1 is inhibited (Bozzaro and Peracino, 2013).
Along with iron, the metabolism and homeostasis of other divalent transition metals – such as divalent manganese, copper, cobalt, and zinc ions – demonstrate similar roles in the resistance of pathogenic infections as part of the host defense mechanism (Bozzaro and Peracino, 2013) (Peracino et al., 2012). Manganese starvation, akin to iron starvation, involves the depletion of manganese from phagosomes through transporter proteins. However, cytosolic copper and zinc influx into the phagosomes and macropinosomes are observed in amoeba upon the ingestion of pathogenic bacteria. This is because the accumulation of copper and zinc in the phagocytic compartments results in the poisoning of the bacteria with high concentrations of divalent copper and zinc, thereby contributing to resistance to bacterial infections through metal poisoning.
The role of reactive oxygen species in cell-autonomous defenses
A primary method of antibacterial host resistance in MPS cells involves reactive oxygen species (ROS). ROS are generated by NADPH oxidase (Dunn et al., 2018). Once recruited to the membrane of the phagosome, NOX2, catalytic subunit of NADPH oxidase (NOX), oxidizes cytosolic NADPH by transferring an electron from this electron-rich compound to oxygen, thus producing a ROS known as superoxide in the phagosome or in the extracellular space. Superoxide then undergoes a series of reactions to produce a variety of other ROS compounds. For example, in one reaction, superoxide can be converted to hydrogen peroxide by superoxide dismutase, and hydrogen peroxide can be subsequently reacted with the ferrous ion in the Fenton reaction to produce hydroxyl radicals.
Due to the oxidative damage caused by ROS, host cells have evolved mechanisms such as encoding and manufacturing enzymes responsible for ROS detoxification, for example, superoxide dismutase, catalase, and peroxiredoxins. Similarly, bacteria and other microbes in the phagosome also express several of the same proteins, such as superoxide dismutase and catalase, to detoxify ROS in the phagocytic compartments. Interestingly, superoxide dismutase requires iron as a cofactor in order to convert superoxide to non-toxic water and hydrogen peroxide (Juttukonda and Skaar, 2015). Therefore, it is hypothesized that the host defense mechanism of iron starvation also prevents the detoxification of ROS by superoxide dismutase, and thereby increases the concentration of ROS in the microbe in the phagosome. This then results in elevated oxidative stress in the bacteria due to an imbalance between the production of free radicals and the ability of the microbe to detoxify them, which accumulates to cause oxidative damage to the microbe. However, it is shown that certain bacteria have evolved to replace iron-superoxide dismutase with manganese-superoxide dismutase, thus enabling them to overcome the iron cofactor restriction imposed by the host cell and remain capable of detoxifying superoxide to non-toxic water and hydrogen peroxide (Murdoch and Skaar, 2022). In addition to inducing oxidative damage to the bacteria in the phagosome through iron restriction, the influx of zinc into the phagosome through protein transporters such as ZNT1 results in zinc accumulation in the phagosome, which has also demonstrated to induce the production of ROS in the phagosome by NADPH oxidase.
Conclusion
Many chemical design solutions exist within the structure of the amoeba that allow it to survive and persist. These design solutions make amoeboid life processes more efficient and answer the problems that the amoebae face in their environments. As explored, these solutions are embedded in the microstructure of the organism; in the composition of the cell surface and cellular organelles that help the organism adapt to different circumstances, and in the complex signaling pathways that allow the organism to sense and respond to its environmental cues.
The high density of mannose-receptors on the cell membrane allows recognition and uptake of yeast cells and other food particles by the amoebae. To survive extreme heat in habitats where wildfires regularly occur, testate amoeba shift towards possessing xenosomic tests whose chemical composition offer better resistance to high temperatures.
Amoebae use chemical reactivity and properties to their advantage, they have developed specialized G-protein coupled receptors on their cell membrane, which allows them to initiate directed movement towards higher concentration gradients of molecules critical to their survival. In particular, Dictyostelium discoideum exhibits an especially fascinating dual chemotactic response, which enables the organism to adapt its behavior based on environmental conditions. When food, like bacteria, is abundant, the cells use GPCRs to move towards folic acid secreted by bacteria, aiding in prey detection and phagocytosis. In contrast, under nutrient scarcity, D. discoideum shifts to a cooperative mode, relying on cAMP specific GCPRs to organize cell aggregation into colonies. This highlights multicellularity as a chemical design solution evolved to ensure the long-term survival of organisms. Moreover, the dual response showcases D. discoideum versatility in adjusting chemotactic behaviors according to nutrient availability and life stage, transitioning from solitary predation to collaborative multicellular development.
Amoebae use chemical reactivity and properties to their advantage – they have developed specialized receptors and other proteins to sense concentration gradients in their environments, direct their motion, and improve their phagocytic abilities to procure nutrients.
In the ongoing evolutionary arms race between the eukaryotic predator and the potentially pathogenic prokaryotic prey, amoebae have also developed microbicidal mechanisms that harness the metabolic pathways of divalent metal ions. Through immunity adaptations such as iron starvation, metal poisoning, and ROS poisoning, amoebae are model organisms for investigating antimicrobial mechanisms that have evolved in phagocytes.
In conclusion, amoebae have ingeniously integrated chemical principles into their cellular structures, evolving resourceful adaptations to enhance their survival and vitality in their environments. The chemical design solutions of amoebae are exemplary manifestations of the amoebae’s ability to harness their resources for optimal advantage as they are confronted with environmental challenges.
References
Allen, P. G., & Dawidowicz, E. A. (1990). Phagocytosis in Acanthamoeba: I. A mannose receptor is responsible for the binding and phagocytosis of yeast. Journal of Cellular Physiology, 145(3), 508-513. https://doi.org/https://doi.org/10.1002/jcp.1041450317
Armynot du Châtelet, E., Bernard, N., Delaine, M., Potdevin, J.-L., & Gilbert, D. (2015). The mineral composition of the tests of ‘testate amoebae’ (Amoebozoa, Arcellinida): The relative importance of grain availability and grain selection. Revue de Micropaléontologie, 58(3), 141-154. https://doi.org/https://doi.org/10.1016/j.revmic.2015.05.001
Bairati, A., & Lehmann, F. E. (1953). Structural and chemical properties of the plasmalemma of Amoeba proteus. Exp Cell Res, 5(1), 220-233. https://doi.org/10.1016/0014-4827(53)90108-6
Baltoumas, F. A., Theodoropoulou, M. C., & Hamodrakas, S. J. (2013). Interactions of the α-subunits of heterotrimeric G-proteins with GPCRs, effectors and RGS proteins: A critical review and analysis of interacting surfaces, conformational shifts, structural diversity and electrostatic potentials. Journal of Structural Biology, 182(3), 209-218. https://doi.org/https://doi.org/10.1016/j.jsb.2013.03.004
Benga, G., (2012). The first discovered water channel protein, later called aquaporin 1: Molecular characteristics, functions and medical implications. Molecular Aspects of Medicine, 33(5-6), 518- 534. https://doi.org/10.1016/j.mam.2012.06.001
Bozzaro, S., Buracco, S., & Peracino, B. (2013). Iron metabolism and resistance to infection by invasive bacteria in the social amoeba Dictyostelium discoideum [Review]. Frontiers in Cellular and Infection Microbiology, 4(SEP), Article Article 50. https://doi.org/10.3389/fcimb.2013.00050
Consalvo, K. M., Rijal, R., Tang, Y., Kirolos, S. A., Smith, M. R., & Gomer, R. H. (2019). Extracellular signaling in Dictyostelium. Int J Dev Biol, 63(8-9-10), 395-405. https://doi.org/10.1387/ijdb.190259rg
Devreotes, P. N., & Zigmond, S. H. (1988). Chemotaxis in Eukaryotic Cells: A Focus on Leukocytes and Dictyostelium. Annual Review of Cell Biology, 4(1), 649-686. https://doi.org/10.1146/annurev.cb.04.110188.003245
Domínguez-Martín, E., Cardenal-Muñoz, E., King, J. S., Soldati, T., Coria, R., & Escalante, R. (2017). Methods to Monitor and Quantify Autophagy in the Social Amoeba Dictyostelium discoideum. Cells, 6(3), 18. https://www.mdpi.com/2073-4409/6/3/18
Du, F., Edwards, K., Shen, Z., Sun, B., De Lozanne, A., Briggs, S., & Firtel, R. A. (2008). Regulation of contractile vacuole formation and activity in Dictyostelium. Embo j, 27(15), 2064-2076. https://doi.org/10.1038/emboj.2008.131
Dunn, J. D., Bosmani, C., Barisch, C., Raykov, L., Lefrançois, L. H., Cardenal-Muñoz, E., López-Jiménez, A. T., & Soldati, T. (2018). Eat prey, live: Dictyostelium discoideum as a model for cell-autonomous defenses [Review]. Frontiers in Immunology, 8(JAN), Article 1906. https://doi.org/10.3389/fimmu.2017.01906
Ferreira, M. D. S., Mendoza, S. R., Gonçalves, D. S., Rodríguez-de la Noval, C., Honorato, L., Nimrichter, L., Ramos, L. F. C., Nogueira, F. C. S., Domont, G. B., Peralta, J. M., & Guimarães, A. J. (2022). Recognition of Cell Wall Mannosylated Components as a Conserved Feature for Fungal Entrance, Adaptation and Survival Within Trophozoites of Acanthamoeba castellanii and Murine Macrophages. Front Cell Infect Microbiol, 12, 858979. https://doi.org/10.3389/fcimb.2022.858979
Fu Man, S., & Casadevall, A. (2018). Divalent Metal Cations Potentiate the Predatory Capacity of Amoeba for Cryptococcus neoformans. Applied and Environmental Microbiology, 84(3), e01717-01717. https://doi.org/10.1128/AEM.01717-17
Hereld, D., & Devreotes, P. N. (2004). Cyclic AMP Receptors of Dictyostelium. In W. J. Lennarz & M. D. Lane (Eds.), Encyclopedia of Biological Chemistry (pp. 488-493). Elsevier. https://doi.org/https://doi.org/10.1016/B0-12-443710-9/00135-6
Heuser, J., Zhu, Q., & Clarke, M. (1993). Proton pumps populate the contractile vacuoles of Dictyostelium amoebae. J Cell Biol, 121(6), 1311-1327. https://doi.org/10.1083/jcb.121.6.1311
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A., Potapenko, A., Bridgland, A., Meyer, C., Kohl, S. A. A., Ballard, A. J., Cowie, A., Romera-Paredes, B., Nikolov, S., Jain, R., Adler, J., Hassabis, D. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873), 583-589. https://doi.org/10.1038/s41586-021-03819-2
Juttukonda, L. J., & Skaar, E. P. (2015). Manganese homeostasis and utilization in pathogenic bacteria. Molecular Microbiology, 97(2), 216-228. https://doi.org/https://doi.org/10.1111/mmi.13034
Kamato, D., Thach, L., Bernard, R., Chan, V., Zheng, W., Kaur, H., Brimble, M., Osman, N., & Little, P. J. (2015). Structure, Function, Pharmacology, and Therapeutic Potential of the G Protein, Gα/q,11. Front Cardiovasc Med, 2, 14. https://doi.org/10.3389/fcvm.2015.00014
Kim, J. Y., Haastert, P. V., & Devreotes, P. N. (1996). Social senses: G-protein-coupled receptor signaling pathways in Dictyostelium discoideum. Chem Biol, 3(4), 239-243. https://doi.org/10.1016/s1074-5521(96)90103-9
Komsic-Buchmann, K., Wöstehoff, L., & Becker, B. (2014). The contractile vacuole as a key regulator of cellular water flow in Chlamydomonas reinhardtii. Eukaryot Cell, 13(11), 1421-1430. https://doi.org/10.1128/ec.00163-14
Korn, E. D., & Wright, P. L. (1973). Macromolecular Composition of an Amoeba Plasma Membrane. Journal of Biological Chemistry, 248(2), 439-447. https://doi.org/https://doi.org/10.1016/S0021-9258(19)44392-5
Latorraca, N. R., Venkatakrishnan, A. J., & Dror, R. O. (2017). GPCR Dynamics: Structures in Motion. Chemical Reviews, 117(1), 139-155. https://doi.org/10.1021/acs.chemrev.6b00177
Liu, Y., Lacal, J., Firtel, R. A., & Kortholt, A. (2018). Connecting G protein signaling to chemoattractant-mediated cell polarity and cytoskeletal reorganization. Small GTPases, 9(4), 360-364. https://doi.org/10.1080/21541248.2016.1235390
Lorch, I. J. (1973). CHAPTER 1 – Some Historical Aspects of Amoeba Studies. In K. W. Jeon (Ed.), The Biology of Amoeba (pp. 1-36). Academic Press. https://doi.org/https://doi.org/10.1016/B978-0-12-384850-5.50007-5
Marcisz, K., Lamentowicz, M., Gałka, M., Colombaroli, D., Adolf, C., & Tinner, W. (2019). Responses of vegetation and testate amoeba trait composition to fire disturbances in and around a bog in central European lowlands (northern Poland). Quaternary Science Reviews, 208, 129-139. https://doi.org/https://doi.org/10.1016/j.quascirev.2019.02.003
Murdoch, C. C., & Skaar, E. P. (2022). Nutritional immunity: the battle for nutrient metals at the host–pathogen interface. Nature Reviews Microbiology, 20(11), 657-670. https://doi.org/10.1038/s41579-022-00745-6
Pan, M., Neilson, M. P., Grunfeld, A. M., Cruz, P., Wen, X., Insall, R. H., & Jin, T. (2018). A G-protein-coupled chemoattractant receptor recognizes lipopolysaccharide for bacterial phagocytosis. PLoS Biol, 16(5), e2005754. https://doi.org/10.1371/journal.pbio.2005754
Pan, M., Xu, X., Chen, Y., & Jin, T. (2016). Identification of a Chemoattractant G-Protein-Coupled Receptor for Folic Acid that Controls Both Chemotaxis and Phagocytosis. Dev Cell, 36(4), 428-439. https://doi.org/10.1016/j.devcel.2016.01.012
Qin, Y., Payne, R., Gu, Y., Mazei, Y., & Wang, Y. (2017). Short-term response of testate amoebae to wildfire. Applied Soil Ecology, 116, 64-69. https://doi.org/https://doi.org/10.1016/j.apsoil.2017.03.018
Schaap, P. (2016). Evolution of developmental signalling in Dictyostelid social amoebas. Curr Opin Genet Dev, 39, 29-34. https://doi.org/10.1016/j.gde.2016.05.014
Schiöth, H. B., & Fredriksson, R. (2005). The GRAFS classification system of G-protein coupled receptors in comparative perspective. General and Comparative Endocrinology, 142(1), 94-101. https://doi.org/https://doi.org/10.1016/j.ygcen.2004.12.018
Shen, J., Ye, R., Romanies, A., Roy, A., Chen, F., Ren, C., Liu, Z., & Zeng, H. (2020) Journal of the American Chemical Society 142 (22), 10050-10058. DOI: 10.1021/jacs.0c02013
Syrovatkina, V., Alegre, K. O., Dey, R., & Huang, X. Y. (2016). Regulation, Signaling, and Physiological Functions of G-Proteins. J Mol Biol, 428(19), 3850-3868. https://doi.org/10.1016/j.jmb.2016.08.002
Xu, X., Pan, M., & Jin, T. (2021). How Phagocytes Acquired the Capability of Hunting and Removing Pathogens From a Human Body: Lessons Learned From Chemotaxis and Phagocytosis of Dictyostelium discoideum (Review). Front Cell Dev Biol, 9, 724940. https://doi.org/10.3389/fcell.2021.724940