Abstract
The use of toxins and venoms to gain control over a limited number of resources is often observed in competitive relationships between organisms. Antagonistic interactions such as allelopathy, exploitative and interference competition significantly contributing to dynamics of coexistence and coevolution of the involved species. Toxin-producing phytoplankton exude allelopathic compounds that inhibit the growth of their competitors while also providing a means for many large populations of phytoplankton species to coexist on a limited number of resources. The bacteria E. Coli fight for the competitive edge in intraspecies interactions by the development and use of colicin, an extremely effective toxin that kills cells upon absorption of a single toxic molecule. A step away from microorganisms, the fire ant S. invicta uses their potent topical insecticide venom to engage in chemical warfare with other invading ant species. Similarly, sea anemones compete over territory, administering their weaponized venom using nematocysts. Different species will show a remarkable variety of chemical mechanisms and toxin nature. Analysis of these various mechanisms allows for a better understanding of the dynamics and balance of coexistence, as well as resistance towards harmful toxins and venoms.
Allelopathy in Phytoplankton
Chemically Mediated Competitive Inhibition—Allelopathy
In the world of microorganisms, populations are plentiful, and resources are in high demand, resulting in heated competition between species. Many microorganisms have developed chemical mechanisms in order to grasp the advantage over their competitors and compete for limited available resources. One such mechanism is allelopathy—a form of interference competition commonly used in harmful algae bloom (HAB) dynamics, such as the red tide dinoflagellate, Karenia brevis. Allelopathy often involves the release of chemical compounds by the allelopathic species, and can inhibit growth and osmoregulation, compromise photosynthetic processes, increase oxidative stress, and cause membrane integrity loss of competing species (Poulin et al., 2017). This process is chemically mediated and can result in fascinating interactions and population fluctuations in phytoplankton blooms.
Allelopathic Compound Structure in K. brevis
Notorious for causing the Red Tide, K. brevis is a marine dinoflagellate commonly known for its brilliant scarlet algae blooms (Fig. 1). The harmful effects of these blooms—namely neurotoxic shellfish poisoning and fish kills— are a result of the production and release of a family of sodium channel agonists called brevetoxins (Poulin et al., 2017).

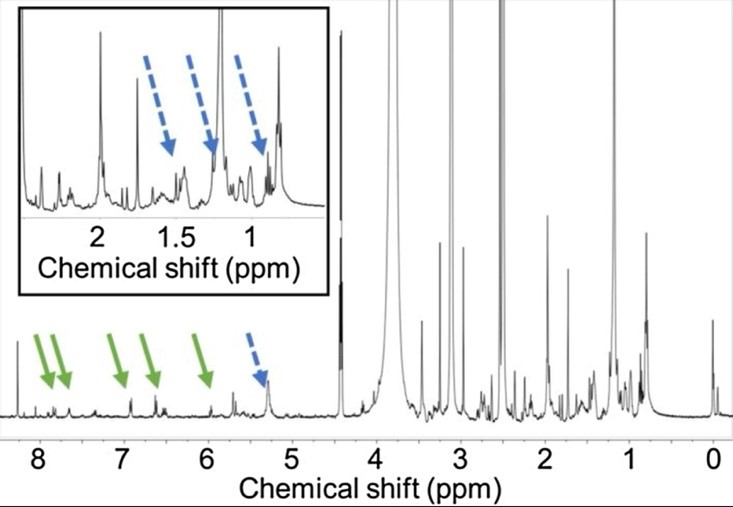
Contrary to expectations, brevetoxins do not have an allelopathic effect on the phytoplankton competitors of K. brevis, such as Asterionellopsis glacialis. A study on the biological variability in allelopathic potency of K. brevis was conducted using A. glacialis growth fluctuations to measure potency and nuclear magnetic resonance spectroscopy (NMR) to identify the key features found in the allelopathic compounds and distinguish them from non-allelopathic compounds (Poulin et al., 2017). It was found that relative to less or non-allelopathic compounds, strongly allelopathic compounds contained enhanced concentrations of fatty acid-derived lipids and aromatic and other polyunsaturated compounds (Fig. 2).
Allelopathy in Phytoplankton Species
Plankton dynamics are mediated strongly by toxin-allelopathy, which can have varying effects on the growth of other micro-algae. It is an important chemical-signalling process and is commonly seen in planktonic or benthic micro-algae, toxic dinoflagellates, and macroalgae which will produce noxious smells. The liberation of allelopathic chemicals gives the toxin-producing species a significant advantage over others in competition for limited resources. However, other than the negative allelopathic effects inhibiting competitors mentioned earlier, in a mixed-species environment, the allelopathic species may exhibit passive mutualisms towards a weaker competitive species (Roy & Chattopadhyay, 2007). Allelopathy is therefore a strong player in aquatic community structure and can greatly alter patterns in species dominance. This will be explored through the dynamic between the three phytoplankton species, Coscinodiscus, Biddulphia, and Chaetoceros.
Phytoplankton—An Exemption to the Competitive Exclusion Principle
A common trend seen in nature to ensure coexistence is the limitation of competing species to prevent them from exceeding a limited number of distinct resources. This trend is called the Competitive Exclusion Principle (Roy & Chattopadhyay, 2007). Remarkable cases have been observed where the number of species coexisting in equilibrium exceed the number of limiting resources. Such cases occur only with the involvement of special mechanisms. Marine phytoplankton communities are a prime example of this situation. Huge populations of many different species are seen in natural water living together on a seemingly limited variety of resources. The reason as to why this occurrence is possible is due to the chemical mechanism of allelopathy.
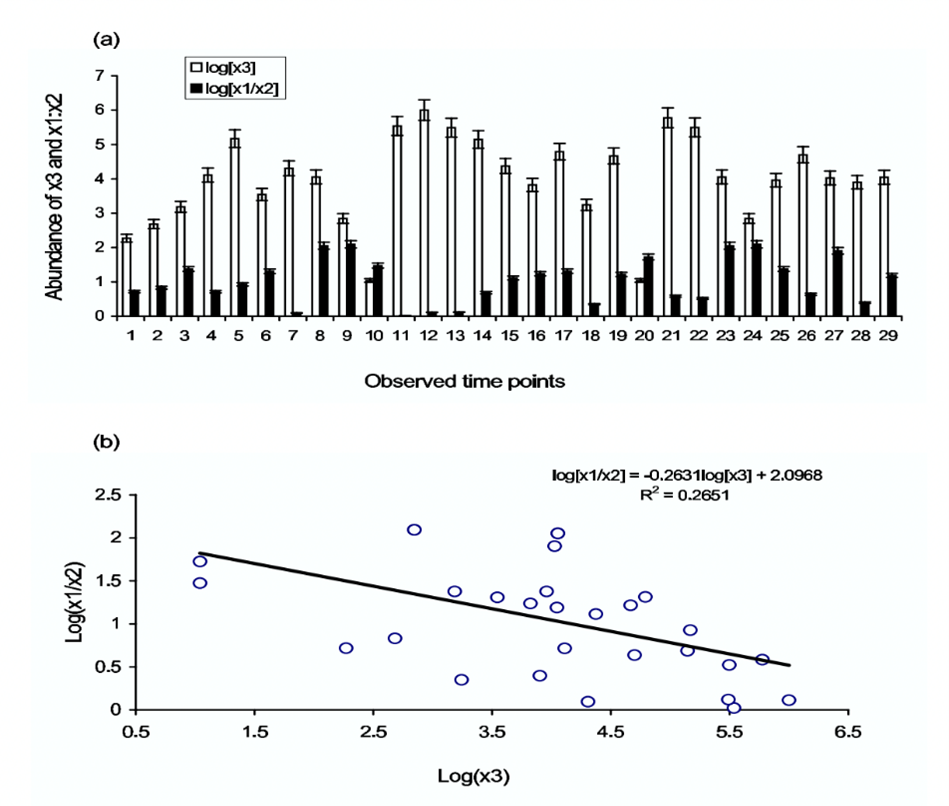
Allelopathic dynamics in phytoplankton revolve around the production of toxic or allelopathic compounds liberated by the toxin-producing phytoplankton (TPP), and how they affect their non-toxic competitors. An example of this relationship is observed between the two non-toxic phytoplankton species, Coscinodiscus and Biddulphia, and the toxin-producing phytoplankton species, Chaetoceros. In a study conducted by Roy and Chattopadhyay (2007), these three species of phytoplankton were put in proximity within an environment containing a limited number of resources, and population growths were recorded. Due to its lower nutrient requirements, Coscinodiscus was the strongest competitor in this dynamic and outcompeted the weaker competitor, Biddulphia, putting them at a disadvantage for resources. However, with the addition of Chaetoceros, the acting TPP in this dynamic, abundance levels of the two non-toxic species underwent interesting fluctuations.
It was observed that when the TPP was sufficiently in abundance, the ratio of abundance between Coscinodiscus (x1) and Biddulphia (x2) decreased by around twenty-five percent (Fig. 3). This result indicated that in the presence of TPP, the competition disadvantage between Coscinodiscus and Biddulphia minimized, which is not favourable for the stronger competitor, Coscinodiscus, but very favourable for the weaker competitor, Biddulphia. Therefore, it was observed that the coexistence of competing phytoplankton species that would otherwise exhibit competitive exclusion on a weaker competitor is determined by the strength of toxin allelopathy and the abundance of TTP (Roy & Chattopadhyay, 2007). Due to this relationship, large phytoplankton communities are allowed to bypass the Competitive Exclusion Principle (Roy & Chattopadhyay, 2007).
Allelopathic Resistance
Under constant exposure to allelopathic compounds by their toxin-producing competitors, certain species of phytoplankton have developed adaptive mechanisms to reduce the allelopathic effects they experience. Consider the harmful algae species, K. brevis, and one of its model competitors, S. costatum. Over their long history of competition, K. brevis’s bloom allelopathy has varied significantly due to S. costatum building a resistance to the growth-inhibiting toxins that the bloom exudes. Not only that, but a study on these two species has observed that the toxin-producing phytoplankton’s allelopathic effects decrease when exposed to S. costatum cells (Prince et al., 2008). There are several possible mechanisms that S. costatum can implicate to undermine K. brevis’s bloom allelopathy. It may degrade or metabolize the toxic compounds released by its competitor, produce defensive chemical compounds to counteract the physiological damage caused by allelopathy, or indirectly depress the production of toxins by depriving K. brevis of resources, since S. costatum exhibits much higher growth rates and has lower nutrient requirements. Despite S. costatum’s lack of allelopathic compounds, the competition between the two species is not one-sided as they constantly adapt and respond to each other to gain the advantage.
Allelopathy is a form of interference competition often seen in communities of phytoplankton. It has significant influence over power dynamics between competing species and provides them a way to coexist while bypassing competitive exclusion. Bacteria behave similarly, with many species making use of deadly toxins to secure resources for themselves.
Intraspecific Interference Competition in Escherichia coli
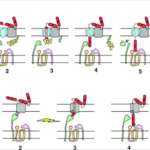
Microorganisms constantly undergo a vicious struggle with competitors in order to secure resources for themselves, each employing various tactics to gain the edge over their competitors. To this end, certain strains of E. coli gain a competitive edge over other members of their species by developing a type of toxin known as colicin. This toxin is extremely effective; the adsorption of a single colicin molecule is enough to result in death of the cell. However, this toxin is highly specific; it targets specific receptors only found on other E. coli cells. In fact, colicin is toxic even to colicinogenic (colicin producing) bacteria. To avoid being killed by their own toxin, these bacteria must generate immunity proteins specific to the type of colicin they produce. Colicin molecules are extremely varied, each inducing cell death through various mechanisms. They do, however, share a few common traits. After being secreted by colicinogenic cells, colicin binds to specific receptors on sensitive bacteria and translocates within the bacterial membrane by using either the Ton-B mechanism or the Tol mechanism (Cascales et al., 2007, Fig. 4).
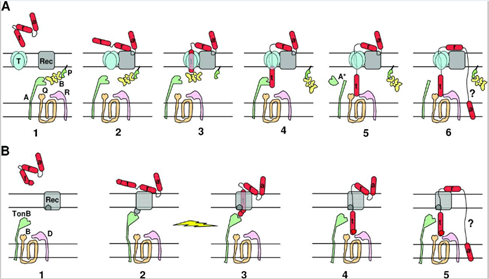
In Figure 4, colicin is depicted in red, with parts of it designated by the letters t, r and a, designating the translocation, reception, and activity domains respectively. The reception domain is responsible for binding to the receptor; the translocation domain allows for translocation of the activity domain through the cell wall; the activity domain triggers biochemical activity leading to cell death. Figure 4A depicts the translocation of colicins with Tol mechanism. In stage 1, the colicin binds to the receptor via the reception domain, followed by translocation of the t domain throughout stages 2 through 5. The mechanism for stage 6 is still poorly understood but does lead to the translocation of the activity domain through the cell wall. Figure 4B depicts the translocation of colicins via the TonB mechanism. In stage 1, the reception domain binds to a specific receptor on the bacterial cell wall, followed by translocation of the t domain through stages 2 to 4. The mechanism for stage 5 is currently poorly understood but results in the translocation of the activity domain through the cell wall.
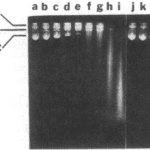
Once inside the cell, colicins can cause cell death through a variety of means. For example, colicin E2 specializes in cell death via degradation of DNA. By acting as an endonuclease, colicin E2 cleaves the phosphodiester bonds which hold DNA together. Through successive cleavage of DNA, DNA is rendered completely unusable, resulting in cell death. A study by K. Schaller and M. Nomura (1976) confirms this; after having completely isolated colicin E2, a test in which DNA samples containing varying concentrations of colicin E2 were analysed through agarose gel electrophoresis showed that high concentrations of colicin E2 would destroy DNA integrity (Fig. 5).

Another type of colicin, colicin E3, induces cell death by cleaving ribosomal RNA; as a result, affected cells become incapable of synthesizing proteins and die off (Braun et al., 1994). Colicins can also induce death by interfering with the production of peptidoglycan, as is the case with colicin M. Since the outer membrane of bacteria is made of this very substance, without the steady production of peptidoglycan, it will degrade, leading to cell lysis (Harkness & Braun, 1988).
Finally, there exists one other way through which colicin induces cellular death. Some types of colicin do not need translocation through the bacterial membrane at all; instead, after binding to a receptor, the colicin turns itself into a K+ channel, imbricated within the bacterial membrane. This leads to the gradual loss of K+ ions through the colicin channel, resulting in depolarization of their membrane. Since bacteria rely on membrane potential to carry out ATP synthesis as well as active transport, colicin molecules effectively disrupt the entire metabolism of the victim cell by inducing depolarization, ultimately leading to cell death (Benarroch & Asally, 2020; Schein et al., 1978).
The various methods through which E. coli provokes cell death might provide inspiration for the creation of alternative antibiotics; this is especially useful, given the rise of antibiotic resistance among bacteria due their widespread usage (Braun et al., 1994).
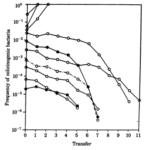
While colicin is an extremely effective toxin, it does not come without disadvantages. The synthesis of the toxin is extremely costly—not only do the bacteria have to synthesize the toxin, but they must also synthesize a corresponding immunity protein in order to avoid being destroyed by their own toxin. Consequently, colicinogenic bacteria have significantly slower growth rates than their non colicinogenic counterparts. A study conducted by Chao and Levin (1981) showed that small populations of colicinogenic bacteria are heavily disadvantaged, as their colicin production is not enough to produce an inhibition zone where other E. coli cannot grow, and their slow growth rates mean their ability to compete for resources is diminished compared to colicin sensitive E. coli. This is readily demonstrated within a liquid culture medium: at low frequencies of colicinogenic bacteria, the strain would eventually die off (Fig. 6). Comparatively, colicinogenic bacteria showed far more success within structured habitats, such as an agar plate, with the producing strain proliferating despite low initial frequencies (Fig. 7). This success can be attributed to the fact that within a structured medium, the inhibition zone produced by a colicinogenic bacterium benefits all the members of its colony (Fig. 8). In contrast, the toxin would only benefit a single individual in a liquid medium.
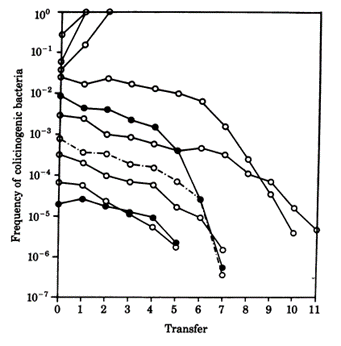

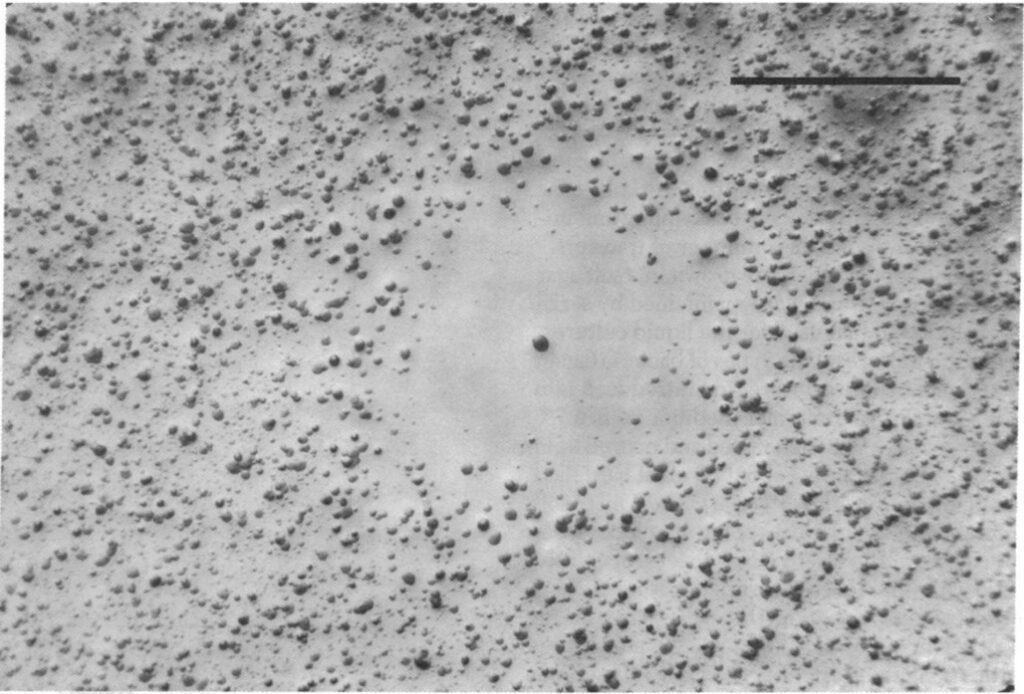
The production of colicin ends up creating a delicate game of rock-paper-scissors within the bacterial ecosystem. As colicin producing bacteria increase in number, certain sensitive bacteria eventually acquire colicin resistant genes, which mutate translocation mechanisms or receptors essential to the action of colicin. As the cost of resistance to colicin is lesser than the cost of generating the toxin and immunity to it, colicin resistant bacteria gain a significant advantage over colicinogenic bacteria. However, colicin resistant bacteria are quickly overtaken by sensitive bacteria, as the latter have faster growth rates since they do not have to generate any resistance genes. And finally, colicin sensitive bacteria lose out to colicinogenic bacteria, as they are quickly destroyed by their toxins (Cascales et al., 2007). Such phenomena are apparent in microorganisms, and can be observed on a larger organismic scale as well—insects can also make use of toxins as a weapon against their competitors, albeit with drastically different mechanisms and effects.
Chemical Warfare and Venom Mechanisms of Ants
Red imported fire ant, S. invicta, is a species belonging to the Formicidae family found in South America and exhibits unique social and competitive behaviors. Over the course of their evolution spurred by competitive dynamics with other species, they developed venom as one of their unique strategies to actively compete and gain an advantage. Such an important tool is useful for the omnivorous fire ant, not only because it provides a means to attack and capture a variety of prey, but also for its role as a defensive measure used by colonies to protect their resources (Touchard, 2016).
Composition, Synthesis and Properties
Generally, the fire ant venom consists of approximately ninety-five percent hydrophobic piperidine alkaloids called solenopsin, and the remaining five percent consists of multiple allergen proteins (Hoffman, 1988).
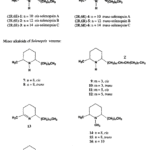
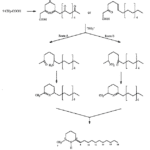
The structure of solenopsin (Fig. 9) is incredibly simple; it consists of a single piperidine ring with a methyl group and a long hydrocarbon chain. Partially due to its simplicity, the chemistry, pharmacology, and toxicology of solenopsin has been studied extensively and several methods of total synthesis have been proposed and tested. However, exact biosynthetic pathways with the biological systems of individual ants remain unclear. A study by Leclercq, Daloze & Braekman (1996) proposes a possible pathway that uses acetate units in linear combination (Fig. 10).
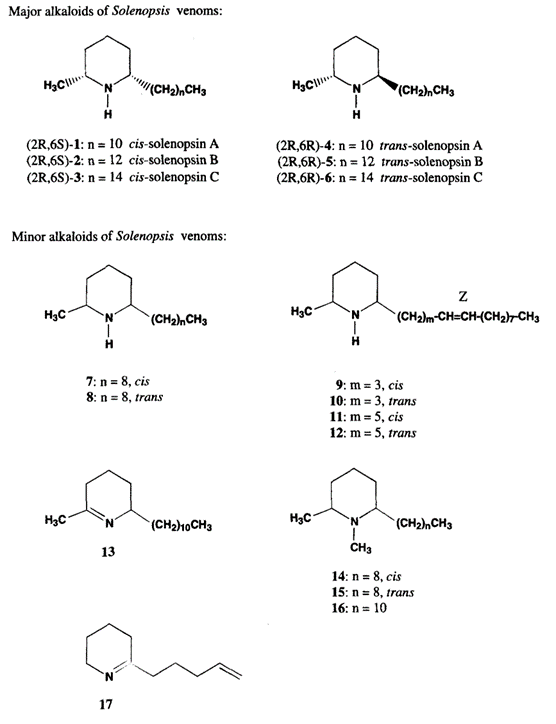
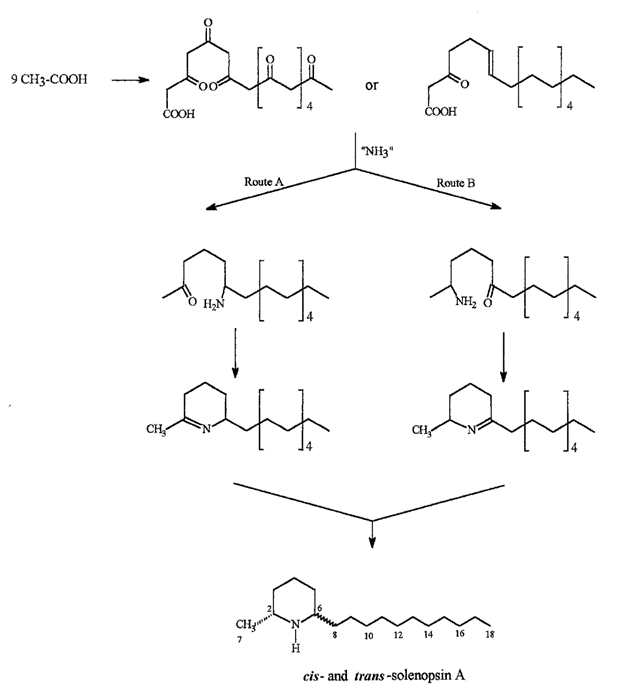
Solenopsin is a potent fire ant venom that exhibits necrotic, antibiotic, and insecticidal properties, enabling the species to use its venom in multiple ways. For example, the insecticidal property can be utilized to kill and capture prey or repel dangerous competitors from the colony. It is also observed that the ants in a colony disinfect the resources and food brought in from the exterior using their own venom due to its notable antimicrobial property (Touchard, 2016).
Other components of the fire ant venom include a number of allergen proteins. In a study by D. Hoffman (1988), four different proteins were separated in the sample of fire ant venom. All four proteins were found to be immunologically unrelated allergens that trigger various degrees of responses in subjects, with none of them being dominant in the sample (Hoffman, 1988). Although the protein components in the venom exist in small proportions compared to solenopsin, together they contribute to a systematic toxic reaction in their victims (dos Santos Pinto et al., 2012).
Chemical Warfare between S. invicta and N. fulva
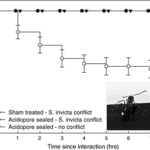
Due to the uniqueness and various beneficial properties of the venom, S. invicta adopts an aggressive competition strategy—invasion. Since the 1980s, records of invasion by S. invicta have been reported, from North and Central America to Asian countries, such as India and China. Along with the invasion, the species brings prominent damage to local agriculture industry and ecosystem (Wang et al., 2019). In the early 2000s, a unique phenomenon was observed in the U.S. Gulf states—the tawny crazy ant (N. fulva), a native competitor of S. invicta with the same South American origin, started to displace S. invicta populations. It was observed that N. fulva gained a significant advantage over S. invicta in this competition, capturing approximately ninety-three percent of the contested common resources (LeBrun et al., 2014). Normally, solenopsin venom is a huge threat to other competitive ant species due to it being a potent insecticide. However, N. fulva developed a unique sequence of behaviours that can successfully detoxify solenopsin, called “detoxification behaviour” (Fig. 11) (LeBrun et al., 2014).
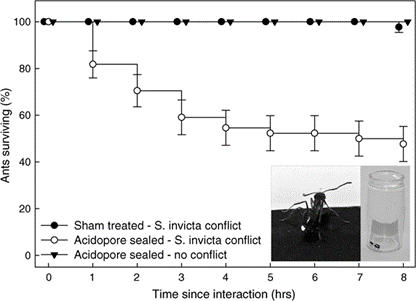
In order to self-detoxify, N.fulva will apply the contents of their acidopore onto the area affected by the S. invicta venom. The study by LeBrun, et al. (2014) hypothesized that one of the chemicals in the fluid of acidopore, formic acid, can detoxify the venom and a series of experiments were conducted. From the results shown in Figure 12 and similar experiments, the study concluded that formic acid is indeed used in the chemical mechanism N. fulva uses to suppress S. invicta and protect themselves from fire ant venom (LeBrun et al., 2014).
Venoms of Sea Anemone Competition
Sea anemones mainly use their venoms to incapacitate and catch prey as well as ward off predators. However, some sea anemones also use their venom to engage in competition over territory with conspecifics (Macrander et al., 2015).
Sea anemones administer their venom using nematocysts, cnidae capsules with tubules capable of penetrating biological tissues, contained in nematocyte cells (Williams, 1991). Sea anemones lack a central venom gland; venoms are produced inside nematocysts and surrounding tissues (Daly, 2017). This allows the organism to deploy a diverse array of toxins specialized for the specific needs of certain tissues (Macrander et al., 2015). The toxins of sea anemones are also unique compared to other venomous animals. Both the generated toxin proteins themselves and their genetic base bear little resemblance to species both outside and inside of the sea anemone’s evolutionary lineage, limiting comparison to better distinguish a toxin’s structure, origin, and role (Daly, 2017).
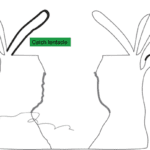
Certain sea anemones have developed specific organs to engage in this competition (Fig. 13). One example includes the catch tentacles of the superfamily Metridioidea (Daly, 2017), which develop from feeding tentacles only after touching another anemone (Williams, 1991). Acrorhagi are another organ used specifically for competition. Acrorhagi are found in some genera of the superfamily Actinioidea (Macrander et al., 2015; Williams, 1991). Located below the base of the tentacles, they resemble hollow spherical growths (Williams, 1991). After repeated tentacle contact with another individual, the acrorhagi begins to expand, and the sea anemone shifts to touch its competitor with its dilated organ. Even if the opponent moves away, parts of the acrorhagi adhere to it (referred to as the “peel”) and inject their nematocysts into the opponent (Williams, 1991). Necrosis occurs in cells surrounding the peels days after injection (Fig. 14).
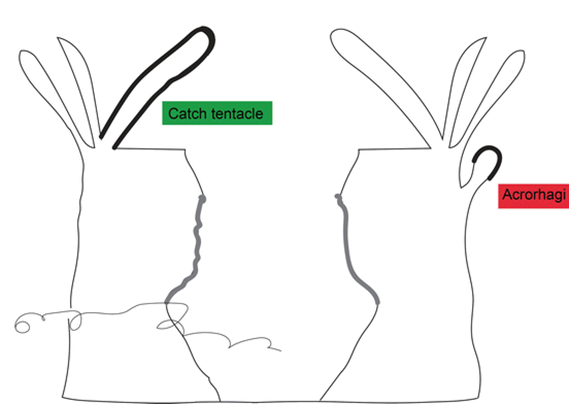
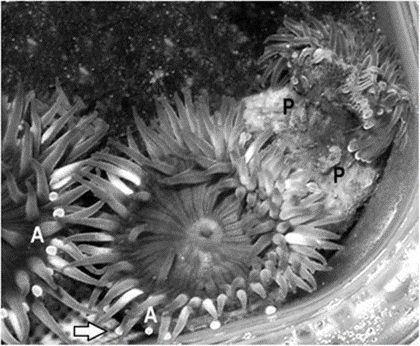
Characterization of Sea Anemone Venoms
Sea Anemones can produce a multitude of venoms with diverse functions. Toxins of similar roles are commonly grouped under one title and are further distinguished into types based on similarities in genetic sequences and target sites (Macrander et al., 2015).
Cytolytic toxins differ widely in structure and mode of action but are joined in their ability to break the membranes of their target cells (Daly, 2017), causing an ionic imbalance that leads to cytolysis (Macrander et al., 2015). They also lack disulfide bonds, unlike the other toxins discussed. Instead, the protein folds due to amino acid residues. Some cytolysins have secondary abilities, such as acting like antihistamines (Type I) or PLA2s (Type III) (Daly, 2017). Type II cytolysins, also called actinoporins, are the most diverse cytolytic toxins and bind to the lipid sphingomyelin (Daly, 2017), However, they are likely not responsible for necrosis from competition as sea anemone cell membranes lack this lipid (Macrander et al., 2015).
Voltage-Gated Channel toxins, or neurotoxins, specifically inhibit the ability of ion channels to control ion flow, critical to several cellular systems such as action-potential generation and propagation by nerve cells (Daly, 2017). Depending on the toxin, it will target either sodium (VGSC) or potassium (VGPC) pumps. Toxins that target VGPCs are more diverse than their VGSC targeting counterparts, likely due to the greater variety of VGPC structures (Macrander et al., 2015). The expression of the various types of both toxins varies across species. Of special note is the type II VGPC toxin, which also acts as a Kunitz-type protease inhibitors, which can generate a multitude of other toxic effects. This jumbles any attempts at characterizing its true function and evolutionary line (Daly, 2017).
Acrorhagins were discovered during the study of acrorhagi. A completely novel toxin, it was originally thought to be used exclusively by sea anemones with acrorhagi for competition (Macrander et al., 2015). However, further research has found similar toxin genes in the genome of Metridium senile, a species unrelated to any with acrorhagi, but does use similar structures for competition (Macrander et al., 2015). Other toxins, such as phospholipase A2s enzymes (PLA2s) and cysteine rich secretory proteins (CRiSP) have been identified due to their surprising resemblance to known animal toxins, such as the venoms of snakes. However, due to the complex venom phenotype of the sea anemone, their role in the diverse library of available venoms is currently unknown (Daly, 2017).
Toxins of Aggressive Sea Anemones
Anthopleura elegantissima are heavily reliant on their acrorhagi to engage in intense competition with members of the same species. A. elegantissima form clonal aggregations of high population densities, resulting in a cramped environment. Acrorhagi, as well as their adhered peels and surrounding necrotic tissue, have been identified in high quantities among individuals along the outskirts of these aggregations (Macrander et al., 2015). From this and other observations, it can be concluded that these animals engage in frequent hostile interactions, resulting in A. elegantissima being considered an “aggressive” species (Macrander et al., 2015). Due to the specific use of acrorhagi by A. elegantissima and the nature of venom generation in sea anemones, it may be possible to identify the specific toxins used by the animal to compete, either through direct study or inspecting toxin-generating genes. A recent attempt to identify these toxins was conducted by Macrander et al. (2015). Their method involved the collection of RNA sequence samples from the surrounding cells of A. elegantissima acrorhagi, both to efficiently construct a complete transcriptome of the species and to determine the relative abundance and expression of certain toxin-generating transcripts. The data was compared to a pool of “non-aggressive” sea anemone genetic data to identify and compare toxin sequences and their abundance. Candidate toxins belonging to each of the groups previously discussed were identified through various techniques, such as BLAST algorithm searches accompanied by the confirmed presence of cysteine amino acid residues characteristic of toxin genes (Macrander et al., 2015).
The results of the study were inconclusive and unexpected. Due to the novelty of the application of the technique and general obscurity sea anemone toxins, many more candidate toxins were identified than previously considered. However, the technique showed promise. Compared to “non-aggressive” species, type II Voltage-Gated Potassium Channel toxin and type I acrorhagin sequences were more abundant in A. elegantissima, possibly indicating the toxins play a relatively greater role in intraspecies competition (Macrander et al., 2015). It was also shown that some toxin transcript levels were elevated during acrorhagi inflation, indicating increased expression and production before the period of injection (Macrander et al., 2015). No sea anemone venom has yet been identified as tissue-specific and any general understandings of the nature and function of venoms used in their competition remains limited (Daly, 2017). However, after continued research as well as improvements to research methods (Macrander et al., 2015), the answer to a venom’s use and evolution may be identifiable.
Conclusion
Mechanisms of toxicity are observed with fascinating variation across organisms—from the microorganisms E. coli bacteria and Chaetoceros phytoplankton, to larger multicellular life forms such as sea anemones and ants. While toxin generation may be costly for the toxin-producing organisms, the competitive advantages it provides greatly outweigh its price of expenditure. For example, S. invicta fire ant’s venom is incredibly versatile; it can be used as both a detoxifier and weapon against other ant species. Sea anemone toxins can provoke a wide variety of lethal effects, such as cell lysis or the inhibition of ion regulation. Additionally, the evolution of venom in organisms promotes biodiversity. As toxin-wielding organisms proliferate, so do toxin-resistant competitors, ensuring a balance within ecosystems. This equilibrium is represented by interactions between colicinogenic, colicin resistant, and non-colinogenic E. coli, which coexist in a delicate balance resembling a game of rock-paper-scissors. Similarly, allelopathic features of the phytoplankton Chaetoceros evens out the playing field for other competing phytoplankton with a large competitive disparity, exhibiting mutualism with the weaker species. This relationship allows for enormous communities of different phytoplankton to coexist on a limited number of resources despite the Competitive Exclusion Theory. These chemical dynamics between and within species allow for deeper insight into competitive relationships and adaptivity in nature.
References
Benarroch, J. M., & Asally, M. (2020). The Microbiologist’s Guide to Membrane Potential Dynamics. Trends in Microbiology, 28(4), 304-314. https://doi.org/10.1016/j.tim.2019.12.008
Braun, V., Pilsl, H., & Groß, P. (1994). Colicins: structures, modes of action, transfer through membranes, and evolution. Archives of Microbiology, 161(3), 199-206. https://doi.org/10.1007/bf00248693
Cascales, E., Buchanan, S. K., Duché, D., Kleanthous, C., Lloubès, R., Postle, K., Riley, M., Slatin, S., & Cavard, D. (2007). Colicin Biology. Microbiology and Molecular Biology Reviews, 71(1), 158-229. https://doi.org/doi:10.1128/MMBR.00036-06
Chao, L., & Levin, B. R. (1981). Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci U S A, 78(10), 6324-6328. https://doi.org/10.1073/pnas.78.10.6324
Daly, M. (2017). Functional and Genetic Diversity of Toxins in Sea Anemones. In P. Gopalakrishnakone & A. Malhotra (Eds.), Evolution of Venomous Animals and Their Toxins (pp. 87-104). Springer Netherlands. https://doi.org/10.1007/978-94-007-6458-3_17
dos Santos Pinto, J. R. A., Fox, E. G. P., Saidemberg, D. M., Santos, L. D., da Silva Menegasso, A. R., Costa-Manso, E., Machado, E. A., Bueno, O. C., & Palma, M. S. (2012). Proteomic View of the Venom from the Fire Ant Solenopsis invicta Buren. Journal of Proteome Research, 11(9), 4643-4653. https://doi.org/10.1021/pr300451g
Godfrey, M. (2016). Red Tide. Microbial Life. https://serc.carleton.edu/microbelife/topics/redtide/general.html
Harkness, R. E., & Braun, V. (1989). Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J Biol Chem, 264(11), 6177-6182.
Hoffman, D. R., Dove, D. E., & Jacobson, R. S. (1988). Allergens in Hymenoptera venom. XX. Isolation of four allergens from imported fire ant (Solenopsis invicta) venom. J Allergy Clin Immunol, 82(5 Pt 1), 818-827. https://doi.org/10.1016/0091-6749(88)90084-x
LeBrun, E. G., Jones, N. T., & Gilbert, L. E. (2014). Chemical Warfare Among Invaders: A Detoxification Interaction Facilitates an Ant Invasion. Science, 343(6174), 1014-1017. https://doi.org/doi:10.1126/science.1245833
Leclercq, S., Braekman, J.-C., Daloze, D., Pasteels, J., & Vander Meer, R. (1996). Biosynthesis of the Solenopsins, Venom Alkaloids of the Fire Ants. Naturwissenschaften, 83, 222-225. https://doi.org/10.1007/BF01143328
Leclercq, S., Daloze, D., & Braekman, J.-C. (1996). Synthesis of the Fire Ant Alkaloids, Solenopsins. A Review. Organic Preparations and Procedures International, 28(5), 499-543. https://doi.org/10.1080/00304949609458571
Macrander, J., Brugler, M. R., & Daly, M. (2015). A RNA-seq approach to identify putative toxins from acrorhagi in aggressive and non-aggressive Anthopleura elegantissima polyps. BMC Genomics, 16(1). https://doi.org/10.1186/s12864-015-1417-4
Poulin, R. X., Poulson-Ellestad, K. L., Roy, J. S., & Kubanek, J. (2018). Variable allelopathy among phytoplankton reflected in red tide metabolome. Harmful Algae, 71, 50-56. https://doi.org/https://doi.org/10.1016/j.hal.2017.12.002
Prince, E. K., Myers, T. L., Naar, J., & Kubanek, J. (2008). Competing phytoplankton undermines allelopathy of a bloom-forming dinoflagellate. Proceedings of the Royal Society B: Biological Sciences, 275(1652), 2733-2741. https://doi.org/doi:10.1098/rspb.2008.0760
Roy, S., & Chattopadhyay, J. (2007). Toxin-allelopathy among phytoplankton species prevents competitive exclusion. Journal of Biological Systems, 15(01), 73-93.
Schaller, K., & Nomura, M. (1976). Colicin E2 is DNA endonuclease. Proc Natl Acad Sci U S A, 73(11), 3989-3993. https://doi.org/10.1073/pnas.73.11.3989
Schein, S. J., Kagan, B. L., & Finkelstein, A. (1978). Colicin K acts by forming voltage-dependent channels in phospholipid bilayer membranes. Nature, 276(5684), 159-163. https://doi.org/10.1038/276159a0
Touchard, A., Aili, S., Fox, E., Escoubas, P., Orivel, J., Nicholson, G., & Dejean, A. (2016). The Biochemical Toxin Arsenal from Ant Venoms. Toxins, 8(1), 30. https://doi.org/10.3390/toxins8010030
Williams, R. B. (1991). Acrorhagi, catch tentacles and sweeper tentacles: a synopsis of ‘aggression’ of actiniarian and scleractinian Cnidaria. In R. B. Williams, P. F. S. Cornelius, R. G. Hughes, & E. A. Robson (Eds.), Coelenterate Biology: Recent Research on Cnidaria and Ctenophora (pp. 539-545). Springer Netherlands. https://doi.org/10.1007/978-94-011-3240-4_76