Ovis canadensis (“Bighorn Sheep,” 2022)
Abstract
Chemistry is the basis of life. Behind every thought, feeling, movement, or biological function, there are a series of chemical reactions occurring. With this said, it comes as no surprise that the chemical composition of animal horns plays a vital role in the mechanical properties, structures, and functions these cranial appendages display. With this in mind, this essay explores the many layers of animal horns, from their chemical composition and genetic code to their applications from a chemical standpoint. Specifically, this paper begins by exploring the synthesis and meticulous structure of a-keratin to explain the incredible mechanical properties displayed by keratinous horns, such as fracture resistance and energy absorbance, which allow them to complete extremely strenuous tasks. In a similar fashion to keratin, every component of an organism is produced via protein synthesis and encoded within DNA. With this said, the genetics and complicated chemistry behind animal horns are subsequently investigated in order to explain the different horn phenotypes expressed and identify species. Once the chemical and genetic principles of horns are established, their applications can be discussed. Namely, this essay dives into isotopic analysis and environmental toxicity biomonitoring using horns, which ultimately show that there are more uses for animal horns than the conventional use of self-defense and mating rights. The molecular and genetic composition of biomaterials can reveal several fascinating aspects about the species it is derived from, and throughout this paper, it is made clear that horns are no exception.
Introduction
Through the selective tendencies of Mother Nature, such as natural selection and incremental evolutionary steps, nature is capable of designing cellular processes, molecular structures, and incredible tools, such as horns, that fit the niches of many organisms and environments. With this said, it is clear that nature is the perfect engineer and their work on animal horns is just one of their many masterpieces. An animal’s horn is a vital tool evolved to contribute to the animal’s survival, dominance and sexual success (Geist & Walther, 1974). Unfortunately, despite them being a crucial trait to an animal’s success in the wild, evolution has not selected for these appendages to have the ability to regenerate or to self-repair. As valuable as these cranial appendages are, the horns designated to an animal from the moment they are conceived, are the horns they will have for their entire life. Thus, any damage to an animal’s horn will be permanent (Modell, 1969). To this degree, such appendages need to be made of durable materials to withstand the unpredictable and wild environment of the animal kingdom. The molecularly strong chemical composition of animal horns ensures these requirements are met. The following essay explores the chemical and genetic makeup of these appendages and the abnormalities and diseases that occur when a mutation arises in the genetic code specifying horn shape and phenotype. Furthermore, the importance of genetic profiling to help identify species, and gain a further understanding of the chemistry of horns is also discussed to help substantiate current applications of animal horns.
Chemical Composition of Animal Horns
True horns are usually found within multiple families of ungulates. For instance, the Rhinocerotidae (rhinoceros) (Figure 1), the Bovidae (cattle, sheep, goats) (Figure 2), and the Antilocapridae (pronghorn antelope) (Figure 3) are all examples of families which contain true-horned species (Modell, 1969). However, some horns can be seen outside these families, such as in reptiles and invertebrates. For generality, this paper will concentrate on horns of ungulates to dive deep into its chemical analysis and applications but understand that different horn chemistry can exist in species outside of ungulates.

Fig. 1 Rhinoceros – example of an animal in the Rhinocerotidae family (Modell,1969).

Fig. 2 Kudu Ram – example of an animal in the Bovidae family (Modell,1969).

Fig. 3 Pronghorn Antelope – example of an animal in the Antilocapridae family (Modell,1969).
Structure and Chemical Analysis of Ungulate Horns
The root meaning of keratin from Proto-Indo-Europe signifies “horn” (“Keratin,” 2022). With this said, keratin has been known to be an integral component of horns for many years. Ungulate horns consist almost entirely of keratin, the protein responsible for multiple structural functions in the biological world. For instance, it is found in hair, nails, hooves, scales, claws, and other tough epidermal tissues. These cranial appendages extend from the base of the frontal bone of the skull and consist of two main parts; the first part is an epidermal layer that encapsulates the second part, a bony core (Figure 4) (Modell, 1969). In this case, the epidermal layer, known as the horn sheath, is the keratin-based structure (Modell, 1969). It is known that animal horns are essential to an animal’s survival and do not regenerate or repair in the event of damage (Modell, 1969). The chemical composition of this structure needs to consist of a durable and rigid material, which perfectly describes keratin.

Fig. 4 Structure of animal horns depicts the bony horn core and the layers of the 𝛼-keratinous sheath (Goss, 2012).
Now that a basic understanding of the general structure of ungulate horns has been established, the following section will dive into the chemical analysis of keratin and what makes it the ideal material for horns, starting with its synthesis.
Protein synthesis occurs in two steps: transcription and translation. In short, DNA is composed of many nucleotides, which consist of a deoxyribose sugar, a phosphate, and a nitrogen base, and codes for the amino acids necessary to make all the proteins required for life (Miller, n.d.). These bases consist of either adenine, cytosine, guanine, or thymine, and the sequence in which those different nucleotides are arranged can code for various amino acids. In transcription, the segment of DNA that contains the amino acid sequence needed to produce a specific protein, in this case keratin, is converted into the form of mRNA, which then leave the nucleus, is processed, and read by the ribosomes in the cytoplasm.
After transcription is achieved, translation begins in the cytoplasm. In this step, the ribosome reads the codons (sequence of three RNA nucleotides that code for an amino acid) in the mRNA, and then tRNA (small RNA molecules) brings the encoded amino acids to the ribosome. All the amino acids string together in the correct order to produce a polypeptide which will fold, ultimately determining its function and properties (Miller, n.d).
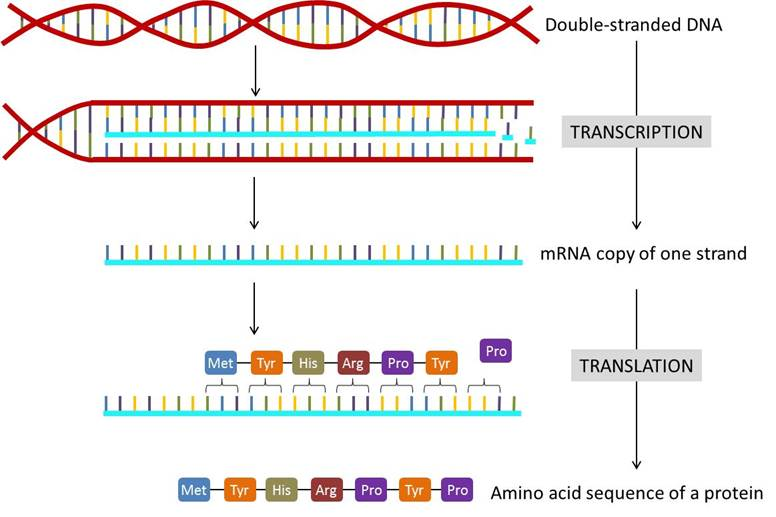
Fig. 5 Transcription and translation summary (Mattaini, n.d.). Segments of DNA are transcribed into mRNA copies, then read in three nucleotide codons which correspond to a sequence of amino acids, which are strung together into a polypeptide.
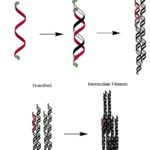
In the case of keratin, its polypeptide chain is rich in cysteine (an amino acid) and it is known to spontaneously self-assemble into bundles of fibers (Zhang et al., 2014). Keratin is classified into two secondary structures alpha-keratin and beta-keratin. Specifically, in animal horns, the sheath is composed of 𝛼-keratin, which folds its polypeptides and forms bundles at a microscopic level that reinforces the entire structure (Zhang et al., 2014). In particular, 𝛼-keratin polypeptides form 𝛼-helices, which then come together with another 𝛼-keratin polypeptide, and twists to form a helical structure known as a coiled-coil. Since keratin is so rich in cysteine, within these dimers, the individual strands are further cross-linked through disulfide bonds involving the cysteine side chains found in the polypeptides (Zhang et al., 2014). These coiled coils can then come together through disulfide bonds with another a-keratin dimer and form a protofilament (Wang et alt., 2016). Once more, these protofilaments bind to produce protofibrils. Ultimately, the protofibrils polymerize to form intermediate filaments (Fig. 6), which make up the lamellae, thin layers of keratin, and the tubules of the horns (Fig. 8). This process gives horns important physical properties, such as reinforced strength through the tubules, and fracture resistance (Wang et al., 2016).
In this way, keratin forms an incredibly tough structure that is among the strongest non-mineralized tissues found in nature (Zhang et al., 2014). Another advantage to the intricately woven 𝛼-keratinous sheath is its ability to remain flexible, while also being tough which allows the structure to absorb energy and limit breakage. Furthermore, due to the coiled folding patterns and disulfide bonds, the structure can flex in many directions and resist breakage (Wang et al., 2016).

Fig. 6 𝛼-keratin structure (Mlpatton, 2017).
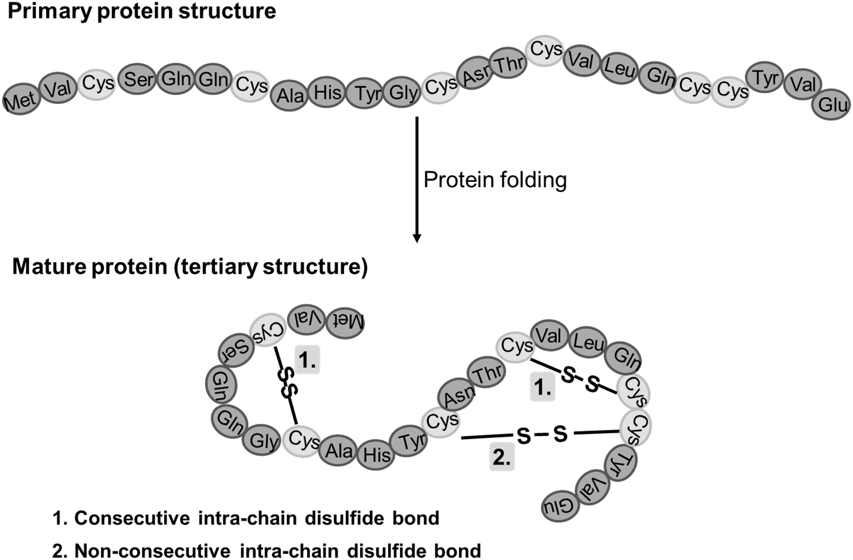
Fig. 7 Example of Disulfide bonds in a mature protein (Creative Proteomics, n.d.).
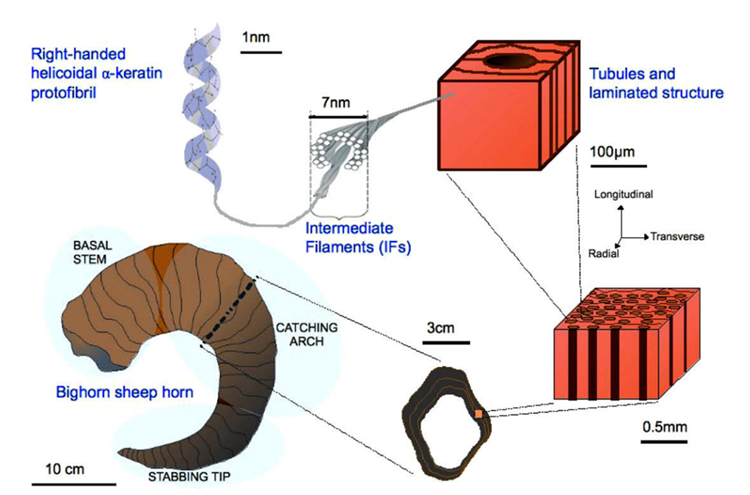
Fig. 8 Keratinous tubules and lamellae structure of the horn sheath (Tombolato et al., 2010).
Elemental Differences Between Horns and Antlers
Horns and antlers are made of tissues which are essential for storing elements such as Calcium (Ca), Phosphorus (P), Magnesium (Mn) and Cadmium (Cd). A study performed by Buddhachet et al. used X-ray fluorescence (XRF) to analyze the chemical composition of horns, antlers and other biological tissues.

Fig. 9 Image of antlers (a) and horns (b) (Selections et al., 2020).
The study analyzed horns and antlers from species of the Bovidae family and Cervidea (deer) family, revealing a difference in 16 out of 19 chemical elements. Chlorine (Cl) was found only in horns, while Vanadium (V), Chromium (Cr), Zirconium (Zr), Silver (Ag), Cadmium (Cd), Tin (Sn) and Antimony (Sb) were present in antlers only. At different concentrations, both antlers and horns shared several elements such as Phosphorus (P), Potassium (k), Calcium (Ca), Titanium (Ti), Manganese (Mn), Iron (Fe) and Sulphur (S). Moreover, significantly higher concentrations of P, K, Ca, Ti, Mn and Fe were detected in antlers, while the horns had a higher concentration of Sulphur, Chlorine and Silicon (Buddhachat et al., 2016).
Chemical composition of horns and antlers
As mentioned above, the results of the study revealed significant traces of Silicon in their elemental composition. In the past, scientists believed silicon had no biological importance and was thus a universal contaminant. However, in recent years it has been made clear that silicon is an important element in collagen synthesis and bone health. Animals consume Si from their environments, whether it be from soil, water or food. Upon excretion, silicon is integrally bound to connective tissues. Collagen is a fundamental and abundant component of connective tissue and bones. As mentioned above, Si is believed to be part of collagen formation as it is part of the gene transcription of type I collagen gene and is also a cofactor in the enzyme, propyl hydroxylase, which is involved in collagen synthesis (Jugdaohsingh, 2007). That being said, collagen is not found in horns. Furthermore, out of the 10 horned subjects, not all their silicon concentrations were the same. This suggests that the accumulation of this element is due to environmental contamination since both soil and water have large amounts of silicon.
Anatomically, horns have an outer keratin tissue layer which covers the appendage which extends from the frontal bone of the animal’s skull. The dermis is continuous with the periosteum and epidermis, creating a protective keratin layer through keratinization. Keratinization requires amino acids, minerals and vitamins such as Zn, Cu, Se and Mn to happen (Buddhachat et al., 2016).

Fig. 10 Anatomy of a developed bovine horn. It can see the keratinous horn sheath surrounds the dermis and the periosteum and extends from the animal’s frontal bone (Rousseau, 2022).
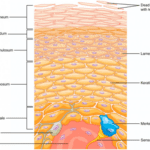
The epidermis is the most exterior layer of a chordate body, which provides protection against invasive microorganisms, moisture loss and provides flexibility to its structure. Additionally, the epidermal layer generates structures such as feathers, claws and of course, horns. Epidermal cells are constantly multiplying in the lower layers of the epidermis, as seen in Figure 11. When new cells are formed, they force more superficial cells to the outer layer of epithelial tissue called the stratum corneum. During apoptosis, these exiled epidermal cells pick up keratin protein products. The cornified skin resulting from this product forms appendages such as horns (“The integument and its derivatives”, 2021).
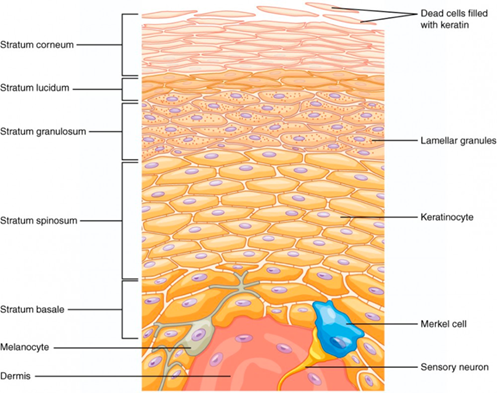
Fig. 11 Layered epidermis. Notice the dead epidermis cells which accumulate keratin within the stratum corneum (“Layers of the Skin | Anatomy and Physiology I”, 2013).
Furthermore, horns contain large traces of Sulphur. One possible explanation for this is its importance in the keratinization process. Keratinization is the formation of a protective tissue which encloses the horn structure. On a molecular level, keratinization involves inducing small changes in tonofibrils to synthesize keratin within the horny cell area (Matoltsy, 1976). As seen in Figure 12, tonofibrils are one of many types of fibers in the cytoplasm of epithelial cells (“Tonofibril”, 2012). Previous research demonstrates that these fibers are bounded by junctions that prevent the fibers from passing from cell to cell. Instead, they terminate at nodes and appear at intercellular bridges (Matoltsy, 1976).

Fig. 12 Schematic image of tonofibrils including tonofilaments, keratin intermediate filaments (“Difference between Tonofibrils and Tonofilaments,” 2020).
Horny cells make up the nonliving component of epithelial tissue. The horny layer is analogues with stratum corneum and is eventually converted into keratin. As seen in Figure 13, densely packed horny cells are recycled products of dead epidermis cells and a product of a combination of tonofibrils (Matoltsy, 1976). Horny cells protect living cells through their many layers of dead cells, which continuously flake off through the process called desquamation. During this process, horny cells go through a series of conformational changes called differentiation. Figure 13 illustrates four types of cell shapes throughout the migration process.

Fig. 13 Densely packed layer of horn cells within the epidermis of skin. (1) Round cells begin to flatten as they rise. (2) Cells become larger and develop prickle-like extensions. This section is known as the prickle-cell layer or stratum spinosum. (3) Cells are completely flat and declared dead (4) the dead horny cells are shed (“The life cycle of a horny cell”, 2003).
Within the cytoskeletal tonofibrils that help in cell adhesion, S-S bond formation occurs and is regarded as the most important element in the keratinization process. These covalent cross-linkages also provide strength and inertness to the tissue (Matoltsy, 1976). Due to its importance in the formation of keratin, sulphur must be abundant within the horn’s elemental library (“The life cycle of a horny cell”, 2003).
Alternatively, antlers share a similar microchemical composition to that of bone. Like osseous tissue, antlers are composed primarily of type I collagen and minerals that assemble into a structural and mechanical scaffold. Unlike horns, antlers have a significant concentration of heavy metals, which may be due to the animal’s diet. For example, deer ingest soil to increase their salt and mineral intake. Finally, antlers contain large amounts of hydroxyapatite, a mineral form of calcium (Ca₅(PO₄)₃), which explains why antlers have a larger concentration of Ca and P (Buddhachat et al., 2016).
As illustrated in Figure 14, deer antlers composition including hydroxyapatite, collagen, non-collagenous proteins and water attributes different bone zones (Picavet & Balligand, 2016). Those components lead to the critical difference between antler and horns; antlers could be shed and regrown yearly, while horns are never shed and continually grow throughout the animal’s lifetime (“Horns versus Antlers”, 2017).
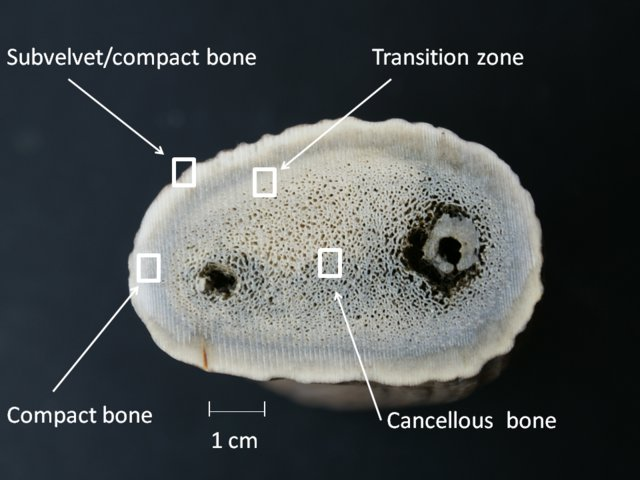
Fig. 14 Cross section of a deer antler (Picavet & Balligand, 2016).
Regenerating Properties amongst Ruminants
Among species that comprise the pecoran families such as giraffids, pronghorns, cervids and bovines, deer antlers are the only completely regenerative organ. Despite sharing several genetic and chemical traits, bovine horns have yet to adopt this regenerative behavior. A study led by Yu Wang deduced that most expressed genes amongst horns and antlers are co-expressed in skin, bone, nerve tissue and testis (Wang et al., 2019). Wang and his team noticed that the rapid cell growth rates in antlers and their gene expression profile was positively correlated to pro-oncogenes and tumor suppressing genes, indicating some sort of relationship between appendage regeneration and cancer resistance (Wang et al., 2019). After gathering the gene expression profiles of several ruminant headgear, scientists believe these appendages originate from neural crest stem cells and chondrogenic neural lineages (Wang et al., 2019). That being said, the antler being an outlier reveals that regenerative properties may exploit oncogenesis pathways. The figure below (Figure 15) reveals the different types of appendages of ruminant and their composition and regenerative status. Additionally, the figure reveals that antlers have a composition that is closer to osteocarcinoma rather than normal bone.

Fig. 15 Neural crest cellular origin of ruminant headgear and the tight control of rapid antler regeneration and low cancer risk in cervids (Wang et alt., 2019).
Abnormalities, Mutations, and Diseases
As seen in the section above, DNA sequences are the blueprint for the production and expression of proteins. Even the smallest of changes in a DNA sequence can completely alter the structure and function of a protein, which in turn can change physical characteristics and result in genetic disorders (Auerbach, 1976). These changes in genetic code are referred to as mutations, which can be caused by errors in DNA replication, cell division, mutagens, or viral infections (Auerbach, 1976). Mutations can be advantageous, detrimental, or neutral, but regardless of their desirability, they increase genetic diversity and sometimes result in fascinating phenotypes (Auerbach, 1976).
Polycerate (supernumerary horns)
In the case of animal horns, polycerate is one such condition caused by a mutation resulting in an interesting phenotype. In this condition, multiple pairs of horns are displayed among certain Bovidae (Duboule, 2022). Evidence of this incredible phenomena has been around for centuries. It was recorded that a four-horned goat from Bulle, Switzerland was transferred to Marie-Antoinettes’ personal farm in the Palace of Versailles at the Queen’s request (Duboule, 2022). The mystery of the multi-horned goat piqued the curiosity of many visitors and scientists but remained unanswered until recently. These results are not usually seen in the wild, however in certain cases, domestic sheep breeds, that have been genetically selected for many generations, are known to display supernumerary horns (Greyvenstein et alt., 2016).

Fig. 16 Polycerate displayed in Manx Loaghtan Sheep (Lotz, 2021).
In a recent study, geneticists studied the genetic determinants of polycerate by analyzing and comparing numerous genome sequences from Bovidae with four horns and control Bovidae. It was revealed that the gene guilty of this phenomenon is the HOXD1 gene, one of the 39 HOX genes, also known as an architect gene, which design the body plan of an organism during its embryonic development (Greyvenstein et alt., 2016). There are a couple of ways in which the mutation arises; in one case study, it was found that there is a four-nucleotide deletion (nucleotides were missing from the DNA sequence) in a single site on the HOXD1 gene. Another case indicated multiple nucleotide variations along the HOXD1 gene, and yet another shows a translocation (DNA from one chromosome is transferred to another chromosome) and deletion within the HOXD1 gene (Allais-Bonnet et al., 2021). It was concluded that the mutations are different among different species, but generally, the non-expression or reduced expression of the HOXD1 gene was reflected in supernumerary horns (Allais-Bonnet et al., 2021). Specifically, this gene defines where horns can emerge on the animal’s head. When HOXD1 is mutated and fewer proteins are produced, the boundaries of where horns are allowed to grow extend, resulting in the splitting of horn buds during embryonic development and the growth of an additional set of horns (Duboule, 2021).
Fig. 17 HOXD1 Gene (Gene Cards, 2017).
Polledness: The Absence of Horns
Horns are a common feature among members of the Bovidae family. They provide several advantages for their host such as during combat, courting and socializing. Hornless animals, in general, do not face any health restrictions, nevertheless, their absence can be devastating for an individual in terms of protection and popularity. Animals born without a horn are a product of naturally occurring mutations known as polledness. There are several molecular differences and genetic variations which cause these abnormalities, which are explored in detail by a study published by Simon et al. (2022). Previous studies have demonstrated that the genetic causes of this abnormality vary based on the species. Thus, it is necessary to study each species individually. This suggests that the mutation of different genes is independent among different species (Simon et al., 2022).
(a) Polledness in Cattle and Mongolian Yaks:
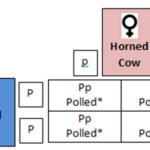
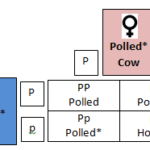
In cattle and Mongolian yaks, the locus which specifies polledness is located on chromosome 1. Polledness is a hereditary dominant trait in which only one copy from a parent triggers the condition. The two figures below illustrate how the allele of the gene is passed down from parents to their offspring.

Fig. 18 Punnett square of the mating of a female horned cow and a male polled cow resulting in an offspring with 100% chance of expressing the polled phenotype (The Genetics of Horned, Polled and Scurred Cattle – Beef Cattle, 2019).
PP = dominant homozygous phenotype
Pp = dominant heterozygous phenotype
pp = recessive homozygous phenotype

Fig. 19 Punnett square of the mating of two polled cows resulting in an offspring with a 75% chance of expressing the polled phenotype and a 25% chance of having horns (The Genetics of Horned, Polled and Scurred Cattle – Beef Cattle, 2019).
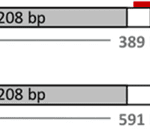
Moreover, there can exist two different P alleles in an individual. Bovidae of Scandinavian and British origin can express the Polled Celtic allele, while those of Holstein Friesian descent express the Polled Freisian allele. The polled Celtic allele, PC, is caused by a small insertion or deletion of base-pairs followed by a duplication of 208 base-pairs found ten base-pairs after the nucleotide sequence that codes for the standard horned phenotype and ends with a six base-pair deletion (Simon et al., 2022).
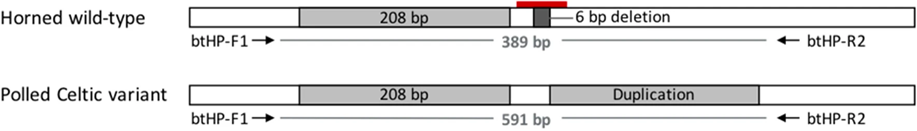
Fig. 20 Contrasted diagrams of chromosome 1 of a horned and polled subject (Schuster et al., 2020).
In contrast, the Polled Freisian allele (PF) is the only polled variant in dairy cattle due to an 80 kilo-base-pair duplication at the locus by three single nucleotide polymorphisms (Schuster et al., 2020). In both cases, the genetic abnormality does not impact protein-coding genes. Instead it affects the noncoding RNA responsible for early horn growth during embryonic development (Simon et al., 2022). Further research reveals that polledness is more likely to occur in the absence of a relaxin/insulin-like family peptide receptor 2, a G protein coupled receptor and protein, forkhead box L2.
Moreover, in yaks there is an additional mutation known as the polled Mongolian allele, PM, which is a 219 base-pair duplication/insertion and seven base-pair deletion and six base-pair insertion 612-bp upstream. The last Polled allele, PG, is found in Nellore cattle and is a 110 kilo-base-pair tandem duplication on chromosome 1 (Simon et al., 2022).
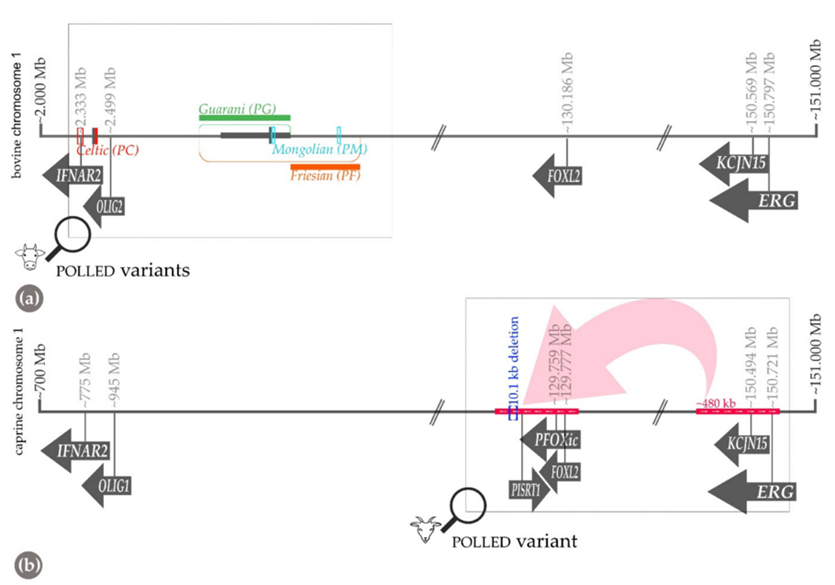
Fig. 21 Schematic diagram of chromosome one for a) bovine and b) goats who express the phenotype for polledness. (a) reveals three locations of alleles PC, PG and PF. Their location along the chromosome is determined by the base-pair nature of their specific mutation (Simon et al., 2022).
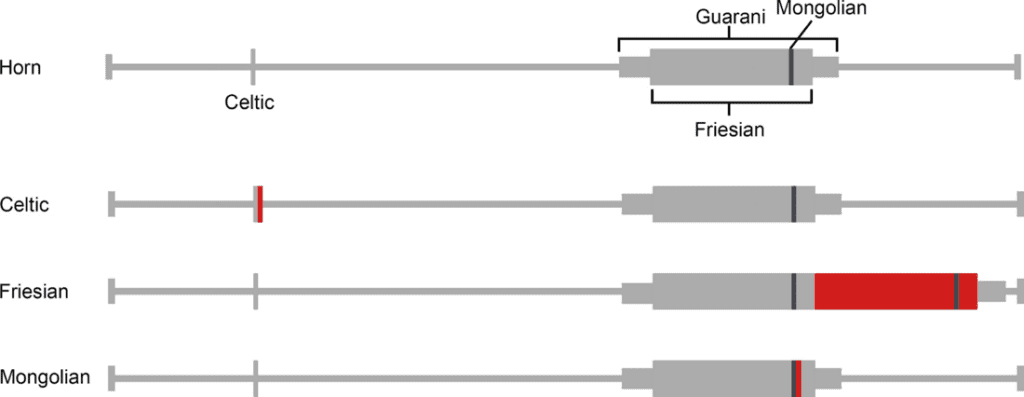
Fig. 22 Schematic representation of Celtic, Fergison and Mongolian mutations. The red rectangle indicates the insertion site of duplicates while the grey represents the duplicated sequence (Aldersey et al., 2020).
Scurs or “Wiggle horns” in cattle:
Scurs are an intermediate phenotype and are often expressed when an animal is heterozygous for a polled allele. This implies that the gene poses one copy of the dominant allele and one copy of the recessive allele. The cause for scurs is still widely unknown; however, researchers believe it adheres to an oligogenic model meaning it is influenced by more than one mutated allele. In addition to a polled heterogynous genotype, a frame-shift variant on chromosome 4 causes the allele to lose its function (Simon et al., 2022).

Fig. 23 Photographs illustrating the scur phenotypes in bovine animals. (A) Brohman cow with fixed scurs (B-C) Bulls with scab-like scurs (Lyons & Randhawa, 2020).
(b) Polledness in Goats:
As with cattle, goats who are polled have a single mutated gene inherited by a parent. The genetic variant is observed to be a 480 kilo-base-pair duplication on chromosome 1 and a deleted part of 10 kilo-base-pair section upstream of chromosome 1.

Fig. 24 Image of a (a) horned goat and (b) heterozygous polled goat. Both (a) and (b) are offspring of a heterozygous polled father (Simon et al., 2022).
Furthermore, researchers have drawn a connection between polledness in goats and recessive intersexuality known as polled intersexuality syndrome (PIS). PIS follows the following genetic blueprint:

Fig. 25 Horn and fertility phenotypes determined by the caprine polled allele in goats (Simon et al., 2022).
Genetics Glossary:
- Gene: Sequence of nucleotides in DNA which serves a specific function. In animals, there are two copies of each gene.
- Genotype: Genetic makeup of an individual
- Locus: location on a chromosome which codes for a specific gene
- Alleles: the DNA sequence at a particular locus
- Heterozygous: A gene with two different alleles
- Homozygous: A gene with two identical alleles
- Genotype: An individual’s entire genetic makeup
- Phenotype: Physical trait associated with a genotype
- Recessive: Masked trait if two dominant alleles are present
- Polled: Absence of horns
- Horned: Presence of horns
- Scurs: Horn tissue attached to the skin instead of the scull
- Smooth Polled: Absence of horns and scars
(The Genetics of Horned, Polled and Scurred Cattle – Beef Cattle. 2019)
Applications of Horn Genetics
The identification of Rhinoceros Species using the DNA from their Horns
An Introduction to Rhinoceroses’:
Rhinoceros horns are a commodity in many parts of Asia. They are used to make art pieces and medical products, making rhinos one of the most endangered species today (Hsieh et al., 2003). To put this into perspective, rhinoceros’ horns are worth more than gold, diamonds and cocaine (Harper et al., 2013). Moreover, identifying rhino products within the market has posed a problem for organizations as their morphological characteristics are still somewhat unknown. This is due to the ambiguous phylogenetic relationship among rhinoceros species. One-way researchers thought to group the species is by geographical areas. The African genera, Ceratotherium and Diceros, and the Asian genus, Dicerorhinus, are placed in a two-horned group, while the Asian genus, Rhinoceros, is the only member in a one-horned group. This hypothesis was supported by analyzing mitochondrial cytochrome b and 12S rRNA gene sequence. The study by Hsing-Mei et al., led to rhinoceroses being characterized into three groups: the two Asian genera, Dicerorhinus and Rhinoceros, respectively and African genera, Ceratotherium and Diceros, as one group (Hsieh et al., 2003).
(a) Cytochrome b gene
Mitochondrial encoded cytochrome b is a protein coding gene which is predicted to be involved in the following activities: metal ion binding, electron transport coupled proton transport, responding to cobalamin, vitamin B12 and glucagon and is in mitochondria (“MT-CYB Mitochondrially Encoded Cytochrome B,” 2022). Cytochrome b is one of eleven elements of the protein complex III, including the bovine cytochrome bc1 protein complex, as in Figure 26. This gene is found in other Mammalian species, such as bovine species and even humans. In fact, it is the only mtDNA found in the respiratory Complex III and mutations in the gene in humans can lead to muscle weakness and exercise intolerance (“Cytochrome B of Complex III,” 2008). This mitochondrial protein is responsible for a single step in oxidative phosphorylation. Oxidative phosphorylation is the process of converting simple sugars into adenosine triphosphate (ATP) and oxygen for cellular use (“MT-CYB Gene,” 2014).

Fig. 26 Bovine cytochrome bc1 protein complex (PDB #1PPJ reproduced with Chimera Software).
(b) Mitochondrial DNA
Mitochondrial DNA (mtDNA) is often used to identify species due to several advantages. mtDNA can resist degradation and has a high copy number. This makes it easier to analyze as cells have roughly 1000 mitochondria with 2 to 10 copies of DNA per organelle (Merheb et al., 2019). Since the experimental procedure involved using powdered horn material, it was necessary to use mitochondrial DNA rather than nuclear DNA due to the degraded nature of the samples.
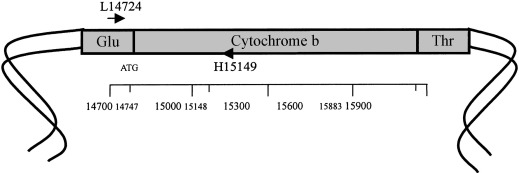
Fig. 27 Cytochrome b loci on mitochondrial DNA (Hsieh et al., 2001).
Experimental Procedure
The study put forth by Hsing-Mei et al. uses powder rhinoceros horns and powdered Holstein cow horn samples. The experiment involved using nested PCR amplification, two primer pairs to amplify the cytochrome b gene and two additional primer pairs to sequence the nested PCR products. The DNA sequences were then aligned using the PileUp program on a GCG computer and a consensus sequence was formed. Their findings show that the most similar sequences are between black and white rhinoceros.

Fig. 28 Example of a trial of partial sentences of cytochrome b aligned using the PileUp program. ‘•’ and ‘N’ represent the same base as the consensus sequence and four nucleotides, respectively (Hsieh et al., 2003).
The largest genetic distance is between Black and Indian rhinoceros. When studying partial fragments of DNA, the results support the hypothesis of a phylogenetic relationship. However, studying full length fragments of cytochrome b yields a different conclusion. With the phenomenon of controversial phylogeny aside, this experiment supports a new method of identifying Rhinoceros DNA using highly degraded samples and will undoubtedly help combat their extinction (Hsieh et al., 2003).
White (Ceratotherium simum) and Black (Diceros bicornis) Rhinoceros DNA Profiling
Similarly, another research group by Halper et al. aimed to genetically profile white and black rhinoceros using a DNA profiling system which included “22 short tandem repeat STR, microsatellite, markers and a gender marker ZFI” (Harper et al., 2013).

Fig. 29 Image of white, Ceratotherium simum, (A) and black, Diceros bicornis, (B) rhinoceros’ horn (Harper et al., 2013).
Background on Microsatellites, PCR and Molecular Markers
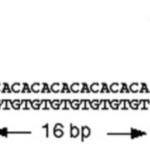
Microsatellites are short sequences of nucleotides which are tandemly aligned and repeated, for example GTGTGTGTGTGT 6 = dinucleotide (12 bp). Scientists use PCR primers to detect microsatellites. These primers are unique to a singular locus and its surrounding base pairs. Thus, one pair of PCR primers will be valid for any individual within a single species (Marwal & Gaur, 2020).
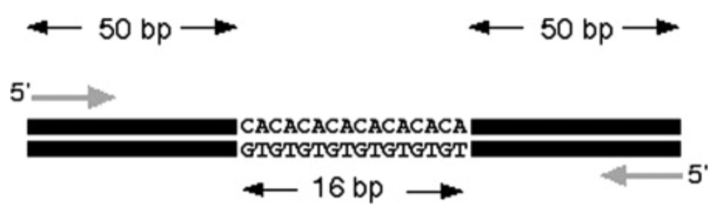
Fig. 30 Identifying microsatellites from DNA using a pair of PCR primers (Marwal & Gaur, 2020).
In the above figure, had there been no microsatellites in the strand, the PCR would be 100 bp in length. In this case the size appears as 116 bp. The difference in predicted length and experimental length reveals the number of repeats in each sample. In this example there are 8 repeats (16 bp / 2 pairs) (Marwal & Gaur, 2020). The study of microsatellites can be useful in understanding species because the combination of base pairs and their length can be unique among individuals, like the patterns that make up fingerprints. As a result, this makes them great phylogenic markers for studying inheritance in families (“Microsatellite,” 2022).
Results of White and Black rhinoceros horn DNA profiling:
The samples collected were inconsistent depending on where they were collected along the horn; however, the center of the horn yields the most consistent and concentrated DNA. The DNA samples collected matched those obtained from other tissue samples such as blood and hair. The study performed by Halper et al. differs from that of Hsing-Mei et al. as they used nuclear DNA extracted from the horn as opposed to mitochondrial DNA. Halper et al. successfully extracted enough adequate nuclear DNA to perform Short Tandem Repeat analysis on both species. After the collection of DNA and genotype analysis, the study compared the expected and observed and compared allele frequencies for expected and actual heterozygosities among the different species. Heterozygosity refers to the level of genetic variation among allele frequencies. A low heterozygosity suggests extensive inbreeding among a particular group, while a high heterozygosity reveals large genetic variation within a population (Philosophy of Biology, 2007). The results of the study revealed an excess in homozygote loci among Black rhinos suggesting a low heterozygosity and large inbreeding coefficient (Gomez-Raya et al., 2015), (Harper et al., 2013).
Chemical Applications of a-Keratinous Horns
Chemical Isotopes in Ungulate Horns
Horned animals are found all over the world in many different environments and conditions. Some follow migratory routes and experience many different habitats. Some species are held captive in zoos or wildlife preservation sites, while others roam free in the wild. While two ungulates of the same species may look similar to the naked eye, many differences in their lifestyles, such as their habitat and feeding habits, can be revealed through the differences in the chemistry of their horns.
Scientists often rely on the analysis of stable isotopes, such as carbon and nitrogen isotopes, to evaluate migration patterns, trophic levels, nutritional habits, and habitat of many animals. This approach depends on foodweb isotopic signatures, which vary based on many biogeochemical processes (the movement of nutrients between biotic and abiotic factors in different environments) (Hobson, 1999). The isotopic signatures from a certain environment are reflected in the tissues of the organisms that live in these conditions (Hobson, 1999). When Carbon, C, and Nitrogen, N, are transferred within food webs, consumers are generally enriched in 13C and 15N isotopes, and by analyzing the ratio of 13C/12C or 15N/14N, it is possible to determine the diet and environment of an organism (Lacumin et al., 2001).
Moreover, different tissues throughout an organism can reveal its diet throughout different periods of its life. For example, analyses of 13C/12C and 15N/14N ratios in bone collagen reflect the isotope signatures of the animal’s average diet within the last 25-30 years of its life. Other tissues, like skin, reveal the diet of the animal throughout a short period of time before its death, and tooth collagen is representative of the isotope signatures during the very beginning of the animal’s life (Lacumin et al., 2001). In some tissues, isotopic analysis can be taken in small segments to provide information on the chronology of isotopic changes (Lacumin et al., 2001). For instance, by taking multiple measurements along a shaft of hair, seasonal differences in the consumption of C3 (plants with 13C/12C ratios ranging from –37 to –21%) and C4 (plants with 13C/12C ratios ranging from –19 to –9.5%) plants can be observed (Lacumin et al., 2001).
Fig. 31 3C/12C Isotopes (Paleontological Research Institute, 2020).

Fig. 32 14N/15N Isotopes (Shelley, 2016).
One study analyzed the outer α-keratinous sheaths of animal horns by testing eight samples of fossilized cattle horn keratin. The goal of the study was to provide a continuous record of isotopes 13C and 15N changes during the keratin formation period (Lacumin et alt., 2001). Seasonal changes affecting vegetation, such as rainfall, temperature and humidity, create variance in an animal’s diet. It was revealed through analyses on small increments in the keratin horn that varying concentrations of carbon and nitrogen isotopes reflect seasonal variance in their diet (Lacumin et al., 2001).
The analysis of 13C was considered to determine the type of plants consumed in the cattle’s life, C3 or C4; C3 plants are typically temperate pastures/vegetation, while C4 are usually tropical pastures or vegetation. On the other hand, analysis of 15N was considered since its values tend to vary based on the trophic level of the organism, and any influence from the climate. For example, a decrease in rainfall causes a 15N enrichment, which can be reflected in horn keratin.
In the results of this experiment, the carbon stable isotope composition of the horn keratin suggests that the cattle grazed a mix of C3 and C4 pasture; C3 or C4 vegetation would dominate depending on the season (Lacumin et al., 2001). The transition period between the seasonal change is reflected through the gradual decrease of the N stable isotope values from the outer to the inner section of the horn. Additionally, the strongly 15N enriched values found in the horns revealed that the cattle resided in an arid environment. Moreover, some of the different specimens exhibited different results in terms of C and N isotope values, which indicate that these cattle were raised in a different environment and, most likely, belonged to a different herd (Lacumin et al., 2001).
Ultimately, it was found that the molecular chemistry of the keratinous horn sheath can record the C and N stable isotope ratios and provide insight into the organism’s diet and local environmental conditions (Lacumin et al., 2001). Throughout the entirety of horned animal’s lives their horns provide them tools for their defense, attacks, and reproductive success. Even after death, the 𝛼 -keratinous horns persist in assisting others, such as researchers, by expressing the story of the horned animal’s life – habitat, diet, lifestyle – through its molecular chemistry.
Horns as a Biomonitoring Model to Test Lead Toxicity
Non-Traditional Monitoring Matrices
There are many cases in which blood, as an analyzable matrix, does not provide enough information to monitor a disease long-term. In the case of lead toxicity, blood provides little information due to its short retention of lead, making it a non-viable monitoring matrix.
Hence, non-traditional monitoring matrices such as analyzing the chemical make-up of horns can be used to bio-monitor specific chemicals such as lead metal. Biomonitoring is defined as the measurement and observation of people’s exposure to environmental contaminants. This analytical technique can provide information about the health impacts certain chemicals have on the body, or potential risks of exposure. It should be noted that the presence of adverse chemicals does not necessarily translate into direct health effects. To establish a direct relationship, other variables and their relationships must be established independently.
Nonetheless, the effectiveness of a biomonitoring approach depends on 3 key parameters: the chemical target, the amount of matrix necessary for the analysis and the detection limit (LOD) of the used analytical technique (Tehrani et al., 2021; Esteban & Castano, 2009).
Various other matrices, such as saliva, breast milk, and semen, are also studied to get an accurate picture of chemicals and proteins in the body, as shown in Figure 32. Of those matrices used, keratinized structures such as horns or hair, offer a stable matrix with numerous advantages. These structures provide easy transport and storage, yield information about short- and long-term exposure, and finally offer a unique temporal exposure pattern by segmental analysis (Esteban & Castano, 2009).
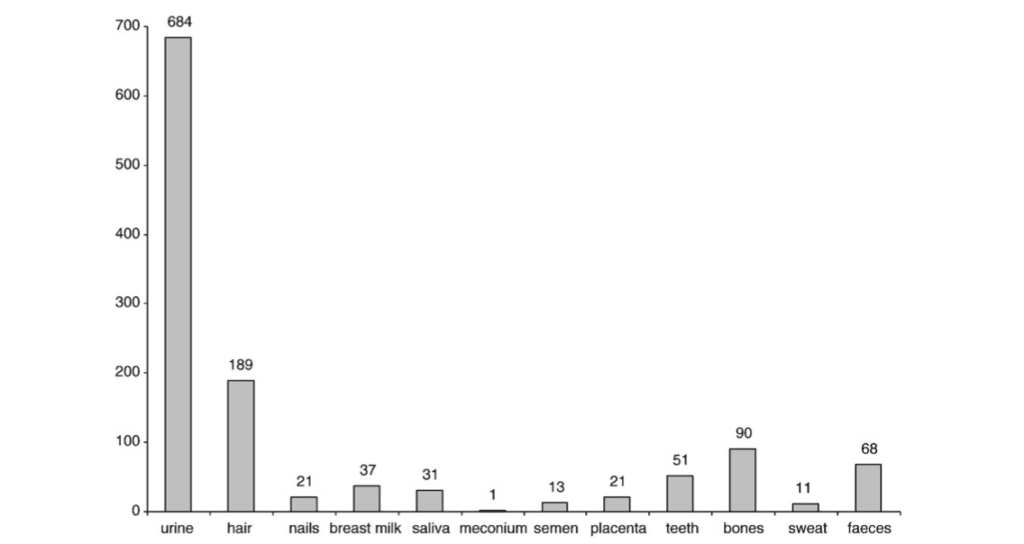
Fig. 32 Distribution bar graph of the number of articles researching the specific matrix, its “respective chemical” as a “monitoring” method. This reveals a significant portion of such matrices include keratinized ones like hair and nails, that bring advantages in being non-invasive and giving important correlations and insights into potential health effects (Esteban & Castano, 2009).
Sampling for Lead Toxicity Exposure Through Keratinized Tissues
Nevertheless, the keratinized structures, whether in horned species or other organisms, offer a strategic biological sampling method in dealing with medical issues on the magnitudes of years or even decades as in the case of lead (Pd) toxicity exposure. This is due to the deposition of lead in the keratinizing tissues as an elimination pathway after the absorption and transportation of lead from the gastrointestinal and respiratory tracts occurs (National Research Council, 1993; Tehrani et al., 2021).
Medically, keratinized tissues offer a critical, years-long record of lead exposures, which if not properly monitored, might develop into real issues if certain thresholds are reached. Researchers use such biological structures in a clinical setting to sample lead (Pb) toxicity levels inside mammalian bodies. Pb toxicity exposure is an environmental risk that accounts for over 1 million deaths and 24 million disability-adjusted life years in 2017. Through its inherently slow residence in the body, reaching magnitudes in decades, blood sampling, as the main biomonitoring action, is insufficient to give an accurate record of lead exposure over time. Testing for blood Pb levels reflects short-term exposure over a period of weeks relative to the actual exposure which could be years to decades.
In turn, non-traditional matrices such as teeth, bones and other keratinous structures, which retain lead traces much longer than blood are being utilized for biomonitoring for Pb toxicity levels in animals (Tehrani et al., 2021).

Fig. 33 Lead (Pb) toxicity is a result of indirect of absorption of lead from the surrounding environment that piles up over decades in various organs/tissues including keratinized structures such as hairs/nails in humans or horns in the case of goats (Ara & Usmani, 2015; Tehrani et al., 2021).
Modeling and Testing Current Biomonitoring Technique for Pb Through Horns
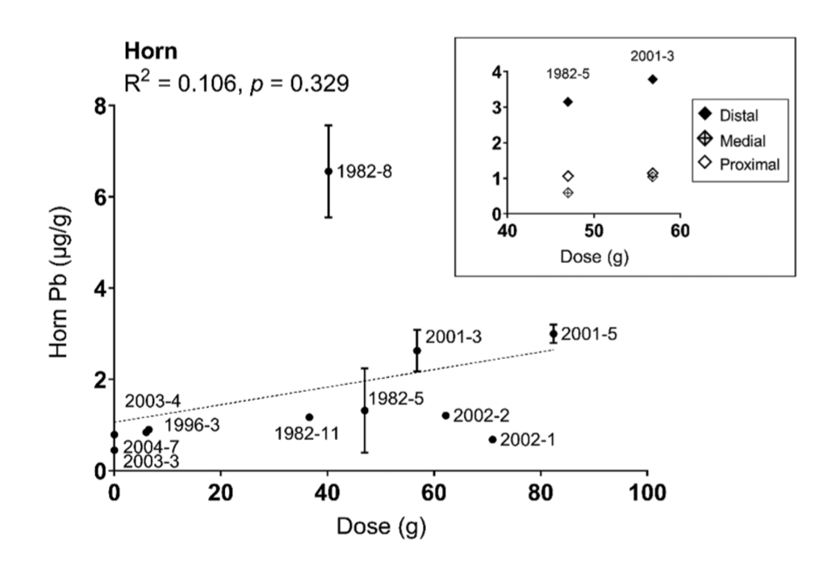
A study by Tehrani et al. (2021) aimed to test several of the previously mentioned bio-monitoring matrices for lead (Pb) by analyzing horns’ Pb levels over decades. The study includes 60 goats and involves interpreting their results in relation to other keratinized structures. Nails and horns, while untraditional in sampling for known diseases, receive lead from blood flow, capillary permeability, and transportation into nearby organs and tissues that increases the rate of absorption. With their long retention rate due to lower growth rate, horns are valuable in looking for a possible correlation between lead exposure and lead toxicity; as shown in Figure 35, this reveals that horn sampling correlates with cumulative levels in bone but peaking at moderate levels and plateauing at the higher doses.

Fig. 34 The variation of observed Pb present in horn sample (µg/g) as a function of dose of lead (g) in 11 horned goats (Tehrani et al., 2021).
Therefore, horns as keratinized matrices are uniquely positioned to allow for the observation, quantification, and analysis of long-term withheld exposure to external toxic sources such as Pb.
Conclusion
Following evolution and natural selection, ungulates and other horned animals have come to possess impressive cranial appendages, which enable them to defend themselves from predators, establish dominance, and earn mating rights. Horn architecture gives the impression that it is simply the shape and mechanical properties that are responsible for these functions; however, its chemical properties are often left out. It is truly the chemical, molecular, and genetic characteristics that form the baseline for all these external functions. The genetic and elemental makeup are the foundation for its impressive and resilient material and shape.
While there exists a vast spectrum of horn shapes and functions, the chemical composition of horns remains consistent among most species. It appears difficult to find a material that will fulfill all the specific niches and functions of the different animal horns; however, as seen throughout this paper, alpha-keratin remains at the forefront of its molecular composition. By further examining its molecular structure, the multi-faceted nature of keratin is revealed, and it becomes clear why it is the ideal material for these cranial appendages. The methodical way that keratin is pieced together allows resilience, flexibility, and strength to withstand all the robust actions required of horns.
Moreover, understanding the possible genetic mutations and phenotype variations, which range from having too many horns, to none, reveals just how important genetic makeup is in ensuring an animal has all the necessary tools for survival. Chemical composition is no different. Comparing elemental compositions to similar structures such as the antler, allows scientists to understand the lack of regenerating capabilities of horns.
Furthermore, the applications of horn chemistry are also discussed in the final section of this paper, where it is revealed that isotopic analysis can identify the diet and environment of horned animals, even after death. Horns with their slower growth rate and retention of internally deposited chemicals offer an alternative matrix instead of blood for the monitoring of long-term exposure to toxic Pb levels.
Ultimately, throughout this paper, it becomes clear that the molecular chemistry of animal horns plays a vital role in their structural integrity, shape, diversity, and research techniques. As observed from the beginning of keratin synthesis from DNA to the firm and unique horn structures that are seen in nature, it is evident that horns are both macroscopically and microscopically strong – a combination that will last a lifetime.
References
Jugdaohsingh, R. (2007). Silicon and bone health. The Journal of Nutrition, Health &
Aging, 11(2), 99–110. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2658806/
The integument and its derivatives. (2021). Zoology.ubc.ca.
https://www.zoology.ubc.ca/~millenoldvertebrate/integument.htm
Layers of the Skin | Anatomy and Physiology I. (2013, April 25). Courses.lumenlearning.com.
https://courses.lumenlearning.com/suny-ap1/chapter/layers-of-the-skin/
The life cycle of a horny cell -Photos & Illustrations. (2003, October 1). Photos & Illustrations.
Retrieved October 25, 2022, from
Marwal, A., & Gaur, R. K. (2020). Molecular markers: tool for genetic analysis. Animal
Biotechnology, 353–372. https://doi.org/10.1016/b978-0-12-811710-1.00016-1
Philosophy of Biology. (2007). Elsevier. https://doi.org/10.1016/b978-0-444-51543
8.x5000-x
CYTOCHROME b OF COMPLEX III; MTCYB. (2008, October 23).
Omim.org. Retrieved November 16, 2022, from https://omim.org/entry/516020
Allais-Bonnet et alt. (2021). Analysis of polycerate mutants reveals the evolutionary co-option of hoxd1
for horn patterning in bovidae. Molecular Biology and Evolution.
https://doi.org/10.1093/molbev/msab021
Auerbach, C. (1976). Mutation research: Problems, results and perspectives. Chapman & Hall.
Duboule, D. (2022, March 23). Médias Médias. University of Geneva. Retrieved October 26, 2022, from https://www.unige.ch/communication/communiques/en/2021/chevres-et-moutons-a-quatre-cornes-le-mystere-resolu
Geist, V., & Walther, F. R. (1974). The behaviour of ungulates and its relation to management: The papers of an international symposium held at the University of Calgary, Alberta, Canada, 2-5 November 1971. International Union for Conservation of Nature and Natural Resources.
Greyvenstein, O. F., Reich, C. M., van Marle-Koster, E., Riley, D. G., & Hayes, B. J. (2016). Polyceraty (multi-horns) in Damara sheep maps to ovine chromosome 2. Animal Genetics, 47(2), 263–266. https://doi.org/10.1111/age.12411
Hobson, K. A. (1999). Tracing origins and migration of wildlife using stable isotopes: A Review. Oecologia, 120(3), 314–326. https://doi.org/10.1007/s004420050865
Keratin – definition, meaning & synonyms (2022). Vocabulary.com. Retrieved October 26, 2022, from https://www.vocabulary.com/dictionary/keratin
Laeumin, P., Bocherens, H., & Louis, C. (2001). Keratin C and N stable isotope ratios of fossil cattle horn from Kerma (Sudan): A record of dietary changes. Italian Journal of Quaternary Sciences, 14(1), 41–46.
Miller, C. (2020, September 1). 5.7 protein synthesis. Go to the cover page of Human Biology. Retrieved October 26, 2022, from https://humanbiology.pressbooks.tru.ca/chapter/5-6-protein-synthesis/#:~:text=Protein%20synthesis%20is%20the%20process,initiation%2C%20elongation%2C%20and%20termination
Modell, W. (1969). Horns and Antlers. Scientific American, 220(4), 114–122. https://doi.org/10.1038/scientificamerican0469-114
Wang, B., Yang, W., McKittrick, J., & Meyers, M. A. (2016). Keratin: Structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Progress in Materials Science, 76, 229–318. https://doi.org/10.1016/j.pmatsci.2015.06.001
Zhang, Z., Ortiz, O., Goyal, R., & Kohn, J. (2014). Biodegradable polymers. Handbook of Polymer Applications in Medicine and Medical Devices, 303–335. https://doi.org/10.1016/b978-0-323-22805-3.00013-x
Hsieh, H.-M., Huang, L.-H., Tsai, L.-C., Kuo, Y.-C., Meng, H.-H., Linacre, A., &
Lee, J. C.-I. (2003). Species identification of rhinoceros horns using the
cytochrome b gene. Forensic Science International, 136(1-3), 1–11.
https://doi.org/10.1016/s0379-0738(03)00251-2
MT-CYB mitochondrially encoded cytochrome b [Homo sapiens (human)] – Gene –
NCBI. (2022, September 22). Www.ncbi.nlm.nih.gov. Retrieved October 24, 2022, from
https://www.ncbi.nlm.nih.gov/gene/4519
MT-CYB gene: MedlinePlus Genetics. (2014, April 1). Medlineplus.gov.
https://medlineplus.gov/genetics/gene/mt-cyb/#:~:text=The%20MT%2DCYB%20gene%20provides
Hsieh, H.-M., Chiang, H.-L., Tsai, L.-C., Lai, S.-Y., Huang, N.-E., Linacre, A., &
Lee, J. C.-I. (2001). Cytochrome b gene for species identification of the
conservation animals. Forensic Science International, 122(1), 7–18.
https://doi.org/10.1016/S0379-0738(01)00403-0
Harper, C. K., Vermeulen, G. J., Clarke, A. B., de Wet, J. I., & Guthrie, A. J. (2013).
Extraction of nuclear DNA from rhinoceros horn and characterization of DNA
profiling systems for white (Ceratotherium simum) and black (Dicerosbicornis) rhinoceros. Forensic
Science International: Genetics, 7(4), 428–433. https://doi.org/10.1016/j.fsigen.2013.04.003
Microsatellite DNA – an Overview. (2021). ScienceDirect.
https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/microsatellite-dna
Gomez-Raya, L., Rodríguez, C., Barragán, C., & Silió, L. (2015). Genomic
inbreeding coefficients based on the distribution of the length of runs of
homozygosity in a closed line of Iberian pigs. Genetics Selection
Evolution, 47(1). https://doi.org/10.1186/s12711-015-0153-1
Hsieh, H.-M., Chiang, H.-L., Tsai, L.-C., Lai, S.-Y., Huang, N.-E., Linacre, A., &
Lee, J. C.-I. (2001). Cytochrome b gene for species identification of the
conservation animals. Forensic Science International, 122(1), 7–18.
https://doi.org/10.1016/S0379-0738(01)00403-0
Simon, R., Drögemüller, C., & Lühken, G. (2022). The Complex and Diverse Genetic Architecture of the Absence of Horns (Polledness) in Domestic Ruminants, including Goats and Sheep. Genes, 13(5), 832. https://doi.org/10.3390/genes13050832
The Genetics of Horned, Polled and Scurred Cattle – Beef Cattle. (n.d.). Beef-Cattle.extension.org. Retrieved October 24, 2022, from https://beef-cattle.extension.org/the-genetics-of-horned-polled-and-scurred-cattle/
Aldersey, J. E., Sonstegard, T. S., Williams, J. L., & Bottema, C. D. K. (2020). Understanding the effects of the bovine POLLED variants. Animal Genetics, 51(2), 166–176. https://doi.org/10.1111/age.12915
The Genetics of Horned, Polled and Scurred Cattle – Beef Cattle. (n.d.). Beef-Cattle.extension.org. Retrieved October 24, 2022, from https://beef-cattle.extension.org/the-genetics-of-horned-polled-and-scurred-cattle/
Buddhachat, K., Klinhom, S., Siengdee, P., Brown, J. L., Nomsiri, R., Kaewmong, P., Thitaram, C., Mahakkanukrauh, P., & Nganvongpanit, K. (2016). Elemental Analysis of Bone, Teeth, Horn and Antler in Different Animal Species Using Non-Invasive Handheld X-Ray Fluorescence. PLOS ONE, 11(5), e0155458. https://doi.org/10.1371/journal.pone.0155458
Selections, N., Canton, & NY. (n.d.). What’s the difference between antlers and horns? NCPR. Retrieved October 24, 2022, from https://www.northcountrypublicradio.org/news/story/19072/20201105/what-apos-s-the-difference-between-antlers-and-horns
Picavet, P. P., & Balligand, M. (2016). Organic and mechanical properties of Cervidae antlers: a review. Veterinary Research Communications, 40(3-4), 141–147. https://doi.org/10.1007/s11259-016-9663-8
Cai, S., Yang, K., Xu, Y., Guan, J., Han, B., Sun, B., Zeng, Y., & Wu, S. (2021). Structure and moisture effect on the mechanical behavior of a natural biocomposite, buffalo horn sheath. Composites Communications, 26, 100748. https://doi.org/10.1016/j.coco.2021.100748
Matoltsy, A. G. (1976). Keratinization. Journal of Investigative Dermatology, 67(1), 20–25. https://doi.org/10.1111/1523-1747.ep12512473
Tonofibril. (2012) Farlex Partner Medical Dictionary. Retrieved October 25 2022 from https://medical-dictionary.thefreedictionary.com/tonofibril
Photos & Illustrations. (n.d.). Photos & Illustrations. Retrieved October 25, 2022, from https://www.carecreations.basf.com/news-media/photos-and-illustrations/2003/10/01/the-life-cycle-of-a-horny-cell#:~:text=The%20outermost%20layer%20of%20the
Difference Between Tonofibrils and Tonofilaments. (2020, November 29). Compare the Difference between Similar Terms. https://www.differencebetween.com/difference-between-tonofibrils-and-tonofilaments/
Tehrani, M. W., Galusha, A. L., Kannan, A., & Parsons, P. J. (2021). Lead uptake into calcified and keratinized compartments of horns from a convenience sample of lead-dosed goats. Journal of Toxicology and Environmental Health, Part A, 84(18), 729-742.
Ara, A., & Usmani, J. A. (2015). Lead toxicity: a review. Interdisciplinary toxicology, 8(2), 55.
National Research Council. (1993). Biologic markers of lead toxicity. In Measuring Lead Exposure in Infants, Children, and Other Sensitive Populations. National Academies Press (US).
Rousseau, M. (2022, January 1). Case 17.3 – Dehorning (J. A. Orsini, N. S. Grenager, & A. de Lahunta, Eds.). ScienceDirect; Academic Press. https://www.sciencedirect.com/science/article/pii/B978032391015600090X
THE INTEGUMENT AND ITS DERIVATIVES. (n.d.). Www.zoology.ubc.ca. https://www.zoology.ubc.ca/~millen/vertebrate/Bio204_Labs/Lab_2__Integument..html
Schuster, F., Aldag, P., Frenzel, A., Hadeler, K.-G., Lucas-Hahn, A., Niemann, H., & Petersen, B. (2020). CRISPR/Cas12a mediated knock-in of the Polled Celtic variant to produce a polled genotype in dairy cattle. Scientific Reports, 10(1). https://doi.org/10.1038/s41598-020-70531-y
Merheb, M., Matar, R., Hodeify, R., Siddiqui, S. S., Vazhappilly, C. G., Marton, J., Azharuddin, S., & AL Zouabi, H. (2019). Mitochondrial DNA, a Powerful Tool to Decipher Ancient Human Civilization from Domestication to Music, and to Uncover Historical Murder Cases. Cells, 8(5), 433. https://doi.org/10.3390/cells8050433
Lect 4. Heterozygosity. (n.d.). Www.uwyo.edu. http://www.uwyo.edu/dbmcd/molmark/lect04/lect4.html
Microsatellite. (2022, May 10). Genome.gov. https://www.genome.gov/genetics-glossary/Microsatellite
Tombolato, L., Novitskaya, E. E., Chen, P. Y., Sheppard, F. A., & McKittrick, J. (2010). Microstructure, elastic properties and deformation mechanisms of horn keratin. Acta biomaterialia, 6(2), 319-330.
Esteban, M., & Castaño, A. (2009). Non-invasive matrices in human biomonitoring: a review. Environment international, 35(2), 438-449.
Mattaini, K. (n.d.) Transcription/Translation [image]. Pressbooks.
https://rwu.pressbooks.pub/bio103/chapter/the-central-dogma-genes-to-traits/\
Goss. (2012) Horn Sheath and Horn Core [diagram]. Mark Witton’s blog.
http://markwitton-com.blogspot.com/2017/
Mlpatton. (2017) a-keratin structure [diagram]. Wikimedia Commons.
https://commons.wikimedia.org/w/index.php?curid=56888235
Creative Proteomics. (n.d.) Disulfide Bonds [diagram]. Creative Proteomics.
https://www.creative-proteomics.com/services/di-sulfide-bond-localization-2.htm
Tombolato et alt. (2010) Lamellae and Tubules [image]. Rhino resource center.
http://www.rhinoresourcecenter.com/pdf_files/126/1263373670.pdf
Lotz, A. (2021) Polyceraty [photograph]. Interweave.
https://www.interweave.com/fiber-nation/next-up-in-fiber-nation-manx-for-the-memories/
Gene Cards. (2017) HOXD1 gene [diagram]. GeneCards: human gene database.
https://www.genecards.org/cgi-bin/carddisp.pl?gene=HOXD1
Paleontological Research Institute. (2020, March 13) Carbon Isotopes [diagram]. Paleontological Research
Institute. https://www.digitalatlasofancientlife.org/learn/paleoecology/biogeochemical-analysis/
Shelley, D. (2016) Nitrogen isotope [diagram]. National Parks Service.
https://www.nps.gov/articles/parkscience32_2_67_shelley_3839.htm
Lee, H., Noh, K., Lee, S. C., Kwon, I. K., Han, D. W., Lee, I. S., & Hwang, Y. S. (2014). Human hair keratin and its-based biomaterials for biomedical applications. Tissue Engineering and Regenerative Medicine, 11(4), 255-265.
Lyons, R., & Randhawa, I. (2020). Improving the Australian Poll Gene Marker Test. https://www.herefordsaustralia.com.au/wp-content/uploads/2022/02/Poll-Gene-Marker-Test.pdf
Wang, Y., Zhang, C., Wang, N., Li, Z., Heller, R., Liu, R., Zhao, Y., Han, J., Pan, X., Zheng, Z., Dai, X., Chen, C., Dou, M., Peng, S., Chen, X., Liu, J., Li, M., Wang, K., Liu, C., & Lin, Z. (2019). Genetic basis of ruminant headgear and rapid antler regeneration. Science, 364(6446), eaav6335. https://doi.org/10.1126/science.aav6335
Bighorn Sheep: Greater vancouver zoo animals. My Website. (2022). Retrieved November 19, 2022, from https://gvzoo.com/animals/bighorn-sheep
Knoll, J. E. (1985). Estimation of the limit of detection in chromatography. Journal of chromatographic science, 23(9), 422-425.
Horns versus Antlers. (2017, August 2). National Parks Service. Retrieved November 19, 2022, from https://www.nps.gov/articles/yell-horns-vs-antlers.htm