ABSTRACT
Cyanobacteria are a phylum of gram-negative bacteria that have been found in a vast range of habitats including Antarctic lakes, hot thermal springs, tropical soil, and even arid climates. These microorganisms have gained attention for their photosynthetic capabilities, long history on Earth, and more recently, eutrophic blooms due to fertilizer runoff and improper waste management that then threatens other organisms. Cyanobacteria produce an abundance of biochemicals such as antioxidants and toxins, while also managing homeostasis through chemical reactions. In this essay, cyanobacteria’s ability to fix nitrogen is explained through the reactions they use, the solutions to O2 degradation of nitrogenases, and the advantages of nitrogen fixation. Then, the CO2 concentrating mechanism that cyanobacteria use to gather CO2 for photosynthesis shows that active uptake transporters are critical for increasing the concentration of HCO3– that then can be converted to CO2 within specialized Rubisco-containing microcompartments. Additionally, the regulation of cyanobacterial oxidative stress which is aggravated by UV radiation includes avoidance mechanisms through migration and microbial mat formation, as well as the repairing of DNA lesions. Defense mechanisms for its oxidative stress are also bolstered by production of many biomolecules such as antioxidants, UV absorbing Mycosporine-like amino acids, and extracellular polysaccharides. Other biomolecule synthesis includes toxins which can be used to block sodium channels, alter cholinergic nerve stimulation, and inhibit serine/threonine-specific protein phosphatase and protein synthesis. These toxins can function as defense mechanisms, responses to stressful environments, signalling compounds, and instigators of allelopathic effects.
INTRODUCTION
According to the cellular endosymbiosis theory, ancient cyanobacteria evolved into chloroplasts in plants and algae (Berman-Frank, Lundgren, & Falkowski, 2003). Cyanobacteria are one of the only groups that can perform both photosynthesis and respiration. In addition, some species can fix nitrogen. It has been proposed that one of the reasons that these organisms are so hardy and persistent is because of the metabolic flexibility that they possess (Sánchez-Baracaldo & Cardona, 2020). Due to their genetic flexibility and unique architecture, they have become exemplary organisms for the photosynthetic synthesis of sugars, polymers, and metabolites. Employing synthetic biology, it is possible to engineer cyanobacteria as solar biofactories that sustainably transform CO2 emissions, sunlight, and water into useful products (Berman-Frank, Lundgren, & Falkowski, 2003).
Cyanobacteria have played a crucial role in the evolution of the Earth by being one of the earliest organisms to provide oxygen to the atmosphere (Mehdizadeh Allaf & Peerhossaini, 2022). The occurrence of oxygenic photosynthesis is unique to cyanobacteria, and various other bacterial groups have developed distinct forms of anoxygenic photosynthesis. Consequently, it is commonly believed that the emergence of oxygenic photosynthesis occurred concurrently with the rise of cyanobacteria. One belief is that ancestral anoxygenic phototrophs gave rise to oxygenic photosynthesis. While cyanobacteria possess two photosystems (PSI and PSII), anoxygenic phototrophs are equipped with either PSI or photosystems like PSII. Some researchers have proposed some sort of ‘protocyanobacteria’ that was formed from 2 anoxygenic photosystems, while others think cyanobacteria evolved through horizontal gene transfer of the photosystems (Sánchez-Baracaldo & Cardona, 2020).
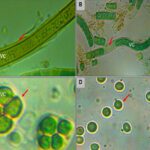
Although cyanobacteria have similar forms and physical structures, they are rich in chemical diversity which allows them to create a palette of various colors (Figure 1) in different environments. Photosynthesis in cyanobacteria follows the same pathway as it does in plants (Vermaas, 2001), however, in addition to the usual chlorophyll-a pigment, and carotenoids found in plants, they also produce other pigments useful for photosynthesis and responsible for the colorful diversity of cyanobacteria (Mehdizadeh Allaf & Peerhossaini, 2022). Cyanobacteria, also known as “blue-green algae,” get their name from the blue-greenish pigment, phycocyanin, which they produce and use to capture light for photosynthesis. But not all cyanobacteria are green; the pigment phycoerythrin, a red protein derived from the phycobiliprotein family, commonly gives a red, brown, orange or even pink color to some cyanobacteria. These bacteria often grow on greenhouse glass or around sinks and drains. As a matter of fact, African flamingos get their pink color from eating cyanobacteria of the genus Spirulina. In aquatic environments, when cyanobacteria are in free floating form, they often form a very dense green cloud that resembles floating paint (Strohmeyer, 2019).

Figure 1. The chemical diversity of cyanobacteria manifested by the palette of colors they can induce in nature (Strohmeyer, 2019).
In this essay, interesting cyanobacterial chemical mechanisms will be explored which gives way to this unique ability of cyanobacteria to survive for long periods of time.
NITROGEN FIXATION
Importance of nitrogen
Nitrogen is the second most prominent element in all living organisms. Approximately 10% of the dry mass of living organisms is composed of nitrogen. Nitrogen is a component of amino acids and is thus essential for the development of proteins, and it is also a component in nucleotides which are the building blocks that make up DNA and RNA. Nitrogen is a component in peptidoglycan cell walls and is required for the synthesis of chlorophylls in photosynthetic organisms. Unfortunately for cells, the highest proportion of available nitrogen in the atmosphere is found as N2, which most organisms cannot use (Stal, 2015).
The stability and near inertness of N2 are attributable to the triple bond between the nitrogen atoms. Although N2 can be chemically converted to NH3, this reaction requires extremely high temperatures and pressures of N2 and H2. It is therefore interesting that bacteria such as Cyanobacteria can perform this conversion under regular environmental conditions (Stal, 2015).
Cyanobacteria nitrogen fixation reaction
Many cyanobacterial species exhibit this special ability to fix nitrogen and are responsible for large amounts of global nitrogen fixation (Latysheva et al., 2012). The process of nitrogen fixation involves the conversion of free nitrogen (N2), a relatively inert gas that is abundant in the atmosphere, into more reactive nitrogen compounds such as ammonia, nitrates, or nitrites. This transformation can occur through either natural or industrial means (Britannica, 2023). In cyanobacteria, nitrogen fixation is performed by nitrogenase enzymes encoded by what are known as nif genes. Nitrogenases exist in three isoforms distinguished by their metallocentres’ metal content: Mo-type (with Mo), V-type (with V), and Fe-type (with Fe). All nitrogen-fixing cyanobacteria have Mo-type, and some also possess V-type, but Fe-type has not been observed in cyanobacteria (Fujita & Uesaka, 2022).
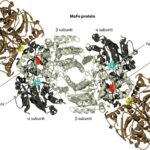
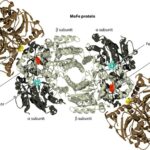
Focusing on Mo-type nitrogenase, as this is the most prominent variety, it is composed of 2 metalloproteins, a dinitrogenase heterotramer, and a dinitrogenase reductase. Dinitrogenase α2β2 heterotetramer has the active site for N2 reduction and its α and β subunits are encoded by the nifD and nifK genes, respectively. Dinitrogenase reductase γ2 homodimer is responsible for transferring high-energy electrons to dinitrogenase and is encoded by the nifH gene (Latysheva et al., 2012). In simpler terms, as seen in Figure 2 below, the dinitrogenase heterotramer is a MoFe protein and the dinitrogenase reductase is a Fe protein (Fujita & Uesaka, 2022).
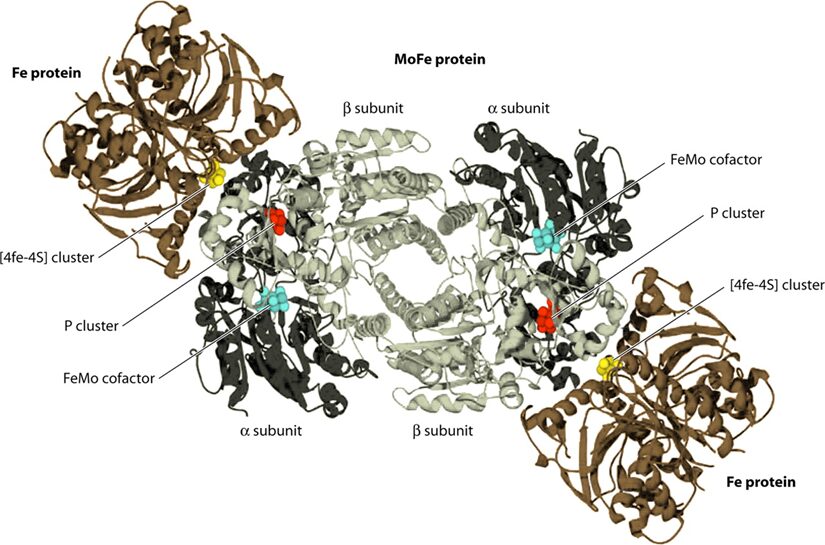
Figure 2. Cyanobacterial nitrogenase, consisting of the Fe protein and the MoFe protein. These components form a temporary complex and contain three metallocenters: Fe4S4 cluster, P-cluster, and FeMo-cofactor, all of which are susceptible to damage by atmospheric or photosynthetically generated oxygen (Bothe, 2010).
The Fe4S4 cluster, produced by a machinery that also creates precursors for the P-cluster and FeMo-co using Cys as a sulphur donor, forms the basis for the P-cluster. Meanwhile, FeMo-co is assembled on a scaffold protein via a complex process involving NifB, homocitrate, molybdenum ion, S-adenosylmethionine, and sulphite, before being inserted into the apo-MoFe protein. Lastly, ferredoxin and ATP, crucial for nitrogenase function, are generated by the photosynthetic electron transport system and the ATPase complex located in the thylakoid membrane of cyanobacterial cells (Fujita & Uesaka, 2022).
These two separable proteins catalyze the nitrogen fixation reaction:
![]()
(Fujita \& Uesaka, 2022)
The nitrogenase reaction (Eq. 1) is intricate, involving at least eight electrons to convert nitrogen into two NH3 molecules via three metal centers, each cycling through various redox states. Additionally, the reaction invariably generates one hydrogen molecule (Fujita & Uesaka, 2022).
Overcoming O2 degradation of Nitrogenases
Cyanobacteria perform photosynthesis as their energy source, which is a problem because oxygen is a product of photosynthesis (Kumar, Mella-Herrera, & Golden, 2010). Nitrogenase, the enzyme for N2 reduction, is very sensitive to oxygen (Stal, 2015). In fact, after exposure to oxygen, the Fe protein’s half-life is approximately 30 seconds, while the MoFe protein’s half-life is around 10 minutes (Stal, 2015). Cyanobacteria have developed various strategies to overcome this sensitivity and still use N2 as a nitrogen source. These strategies include spatial separation, fixing nitrogen at night, and some cyanobacteria even employ a combination of these techniques (Stal, 2015).
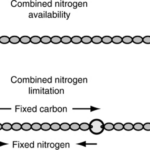
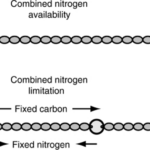
Heterocysts are specialized cells found in filamentous multicellular cyanobacteria which are home to those organisms’ nitrogenases. This solves the problem of the degradation of nitrogenases under aerobic conditions as it separates the nitrogenases from vegetative cells where photosynthesis happens. These cells are differentiated from vegetative cells via cell division and metabolic changes (Bothe, 2010) and are said to be terminally differentiated (Kumar, Mella-Herrera, & Golden, 2010). Heterocysts are distinct from vegetative cells in that they are large and round, have a thicker envelope and less colouration and usually have prominent cyanophycin granules at the poles next to the vegetative cells (Kumar, Mella-Herrera, & Golden, 2010). Cyanophycin is a nitrogen or carbon storage polymer (Björn & Karl, 2018). In some cyanobacteria, such as the filamentous cyanobacterium Anabaena (also Nostoc) sp. strain PCC 7120, cell differentiation depends on nitrogen availability (see Figure 3), and this is therefore an environmental adaptation (Kumar, Mella-Herrera, & Golden, 2010).
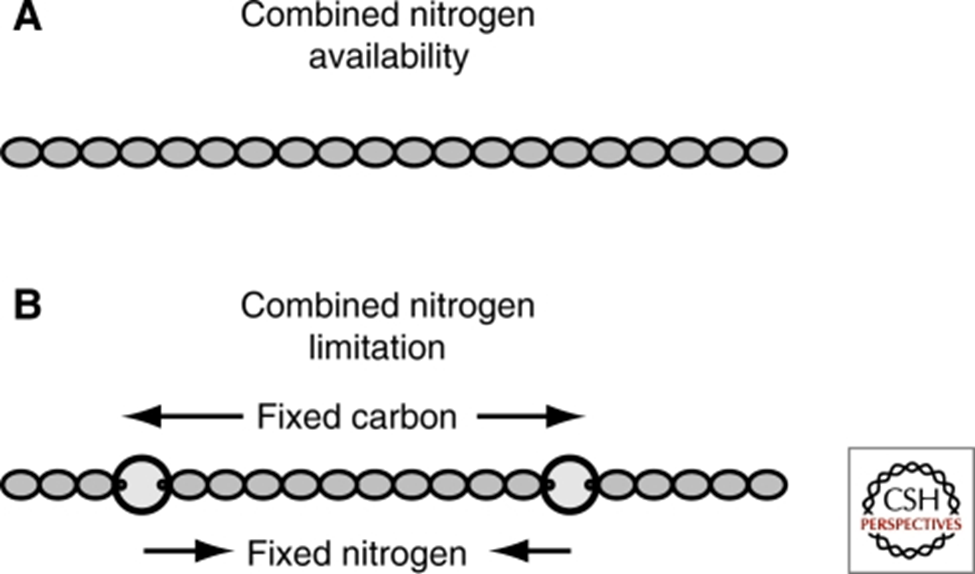
Figure 3. Anabaena PCC 7120 heterocyst development: (A) In nitrogen-rich media, Anabaena PCC 7120 forms filaments of photosynthetic vegetative cells. (B) When nitrogen is scarce, heterocysts form at intervals of 10-20 vegetative cells along filaments (Kumar, Mella-Herrera, & Golden, 2010).
In heterocysts, Photosystem II (PSII), an important component in photosynthesis, undergoes degradation, leaving them incapable of performing the photosynthetic water-splitting reaction. Additionally, they cannot photosynthetically fix CO2 (Bothe, 2010).
In comparison, non-multicellular filamentous, or nonheterocystous cyanobacteria use an internal circadian rhythm, or biological clock in order to protect their nitrogenases (Kumar, Mella-Herrera, & Golden, 2010). In fact, some researchers suppose that it may be the oldest known biological clock (Jabbur & Johnson, 2021). These bacteria separate the two reactions of photosynthesis and nitrogen fixation around the time of day, with photosynthesis performed during the day, and nitrogen fixation at night. Therefore, nitrogenase is not exposed to oxygen which is the by-product of the photosynthesis reaction (Bothe, 2010).
Advantages of Nitrogen Fixation
Nitrogen fixation is crucial for bacteria due to the persistent scarcity of nitrogen in various habitats, particularly in marine environments (Fujita & Uesaka, 2022). Cyanobacteria have evolved to live symbiotically with other organisms. The most common symbiotic relationships are those that nitrogen-fixing cyanobacteria form with fungi, primarily lichens and with the 4 major phylogenetic groups of plants. These relationships provide cyanobacteria with protected environments and access to carbon as an energy source, and in exchange, they provide nitrogen to the other symbiont (Meeks, 1998). In corals, it has been found that cyanobacteria have been incorporated into the coral’s tissues. This benefits the corals, as corals require nitrogen, while the corals also provide an ideal environment with access to carbon via glycerol. Furthermore, coral experiences hypoxia at night, which aids the cyanobacteria in temporal separation to protect their nitrogenases (Lesser et al., 2004).
In summary, nature has evolved cyanobacteria to have nitrogenases which allow them to process nitrogen-containing compounds into nitrogen which can be used in all living organisms. This leads to the creation of symbiotic relationships, in which cyanobacteria can benefit from increased carbon access, protection and better environmental conditions. Furthermore, cyanobacteria have another unique metabolic mechanism when it comes to photosynthesis optimization: CO2 concentration
CO2 CONCENTRATING MECHANISM
Capture of CO2 is one of the key problems that cyanobacteria have faced through evolution and in their current environments. In aquatic environments, CO2 availability and supply rate are often severely limiting. Although the solubility of CO2 in water is higher than that of the other three most abundant gases in the Earth atmosphere (N2, O2, and Ar), CO2 diffusion in water is 104 times slower than in air. The dissolved CO2 reacts with water to form carbonic acid (Kamennaya et al., 2012).
![]()
(Kamennaya et al., 2012)
Subsequently, the carbonic acid dissociates to form bicarbonate (HCO3–) releasing H+ into the solution. A fraction of (HCO3–) further dissociates to form carbonate (CO32-) (Kamennaya et al., 2012).
![]()
![]()
(Kamennaya et al., 2012)
The concentration of H2CO3 at circumneutral pH (5.5 to 7.4) is small, and the dissolved CO2 from the reactions above occurs predominantly as HCO3–, which makes HCO3– to be the main source of CO2 supply. Under this CO2 limiting condition, one might conceive that cyanobacteria need to have a highly efficient thylakoid to sufficiently use the resource to carry out photosynthesis for survival. However, the cyanobacterial Rubisco enzymes (the first enzyme in the Calvin cycle or photosynthetic carbon reduction cycle) possess a low affinity for CO2 (Badger et al., 2006). So how do cyanobacteria survive in such a CO2 limiting environment?
The solution to this puzzle is the development of a CO2 concentrating mechanism (CCM). CCM has two primary functional elements. Firstly, the presence of several inorganic carbon Ci (e.g., CO2, HCO3–) transporters that deliver HCO3– intracellularly. Secondly, the containment of Rubisco in carboxysome protein microbodies within the cell (the sites of CO2 elevation). Rubisco is the first enzyme in the Calvin cycle (or photosynthetic carbon reduction (PCR) cycle) that assimilates CO2 into organic carbon compounds. CCM results in an ensuing accumulation of HCO3– within the cell to ratios as high as 1000-fold with regard to total exogenous Ci levels, irrespective of the type of uptake substrate, CO2 or HCO3– (Badger et al., 2006).
Active Ci Uptake: Ci transporters
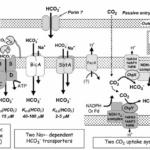
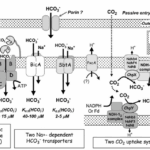
So far, a total of five different uptake systems have been identified (Figure 1), including three HCO3– transporters and two CO2 uptake systems.

(i) BCT1, an inducible high affinity HCO3– transporter encoded by the cmpABCD operon and belonging to the traffic ATPase family ; (ii) SbtA, an inducible, high affinity Na+ – dependent HCO3– transporter ; (iii) BicA, a newly discovered low affinity; ; (iv) NDH-14, a constitutive CO2 uptake system based on a specialized NDH-1 complex that appears to be located on the thylakoid membrane ; and (v) NDH-13, a second CO2 uptake system based on a modified NDH-1 complex that is inducible under Ci limitation (Badger et al., 2006).
As shown in Figure 4 and mentioned above, HCO3– and CO2 are transited into the cell and to the carboxysome. Interestingly, the CO2 in cytosol is first converted to HCO3– and then is transited to the carboxysome to make CO2. This step seems to be unnecessary, but it has valid reasoning. Since the active transporters make the Ci level to be as high as 1000-fold of the exogenous Ci level, Ci are likely to leak out from the cell (Jansson & Northen, 2010). From this perspective, HCO3– is an ion, which means that it is less permeable to lipid membranes than the uncharged CO2 molecule (1000-fold less permeable specifically), which makes it the preferred form of Ci for accumulation, especially where the cytoplasmic pH is 7.8–8.2 (Price et al., 2008).
Despite the low permeability of HCO3– through the lipid layer, Ci still could leak out. To save energy from constantly transiting excess Ci into cell and resulting into the leakage of Ci, the active uptake of Ci needs to be regulated based on the Ci level in the environment. The regulation can be done through changing of the transcription level of the coding genes of the transporters and post-translational regulation. For example, all cyanobacteria so far examined exhibit a basal, low transporter affinity form of the CCM when grown under conditions of Ci sufficiency, indicating that many of the CCM genes coding for Rubisco, carboxysome proteins, and low affinity Ci transporters are essentially under constitutive expression with limited plasticity. However, under Ci-limited conditions, β-cyanobacteria in particular, are able to up-regulate the expression of high affinity Ci transporters such as high affinity CO2 uptake (NDH-I3), SbtA, BCT1, or BicA. The expression of these CO2– responsive genes is controlled by one or two LysR-type transcription regulators known as CcmR (NdhR) or CmpR (Price et al., 2008).
For the post-translational regulation, there is some evidence that activation of HCO3– transporters might involve a redox signal while other evidence suggests that full activation of HCO3– transport might involve phosphorylation via a Ser/Thr protein kinase. It is highly probable that activation involves specific phosphorylation events, but this is not yet proven (Price et al., 2008).
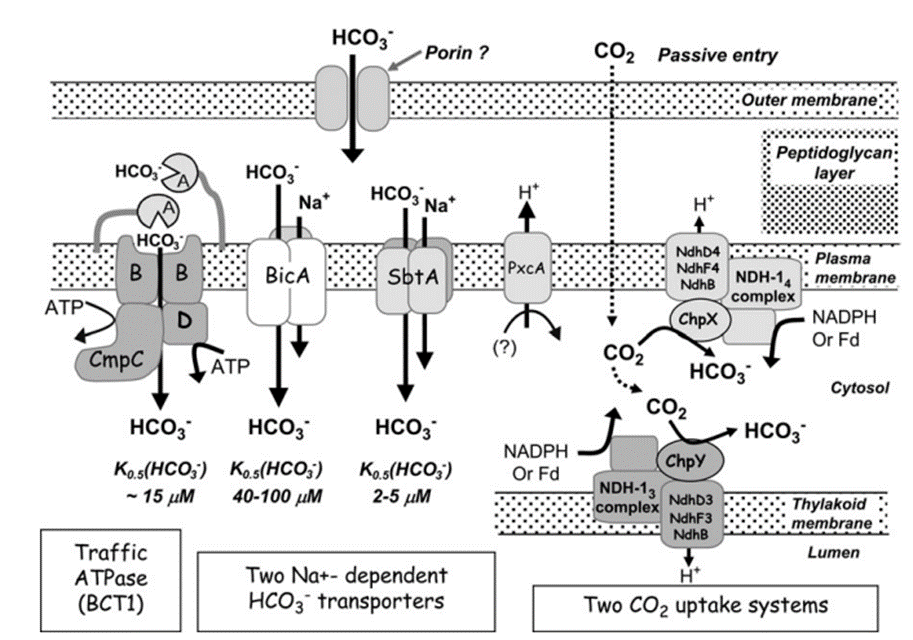
Figure 4. Schema showing the probable structure and membrane location of the five known modes of Ci transport in cyanobacteria. Ancillary components such as porins and PxcA are also shown (Price et al., 2008).
Carboxysome
The accumulated HCO3– is used to generate CO2 within specialized, Rubisco-containing, microcompartments known as carboxysomes. Carboxysomes contain most of the cellular Rubisco content. These structures (90–250 nm in diameter, depending on the cyanobacterial spp.) are proteinaceous, polyhedral bodies featuring tightly packed Rubisco molecules (L8S8) bounded by a thin protein shell of about 3 nm (Badger et al., 2006).
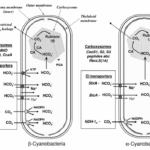
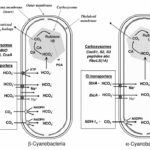
From the viewpoint of Ci acquisition and CO2 fixation, cyanobacteria can be divided in two major groups based on Rubisco and carboxysome lineages. This evolutionary divergence relates to the phylogenetic relationships for Form 1 (L8S8) Rubisco large subunit as either the Form 1A or 1B types. On this basis the two groupings have been referred to as α-cyanobacteria (chiefly oceanic cyanobacteria) for those containing Form 1A Rubisco and β-cyanobacteria (chiefly freshwater strains) with Form 1B Rubisco (Figure 5). Both types of carboxysomes contain a specific carbonic anhydrase (CA) for catalysis of the conversion of HCO3– to CO2. In the case of β-carboxysomes the CA function is carried out by CcaA/IcfA , whilst in α-carboxysomes the shell protein CsoS3 (CsoSCA) is required (Price et al., 2008).
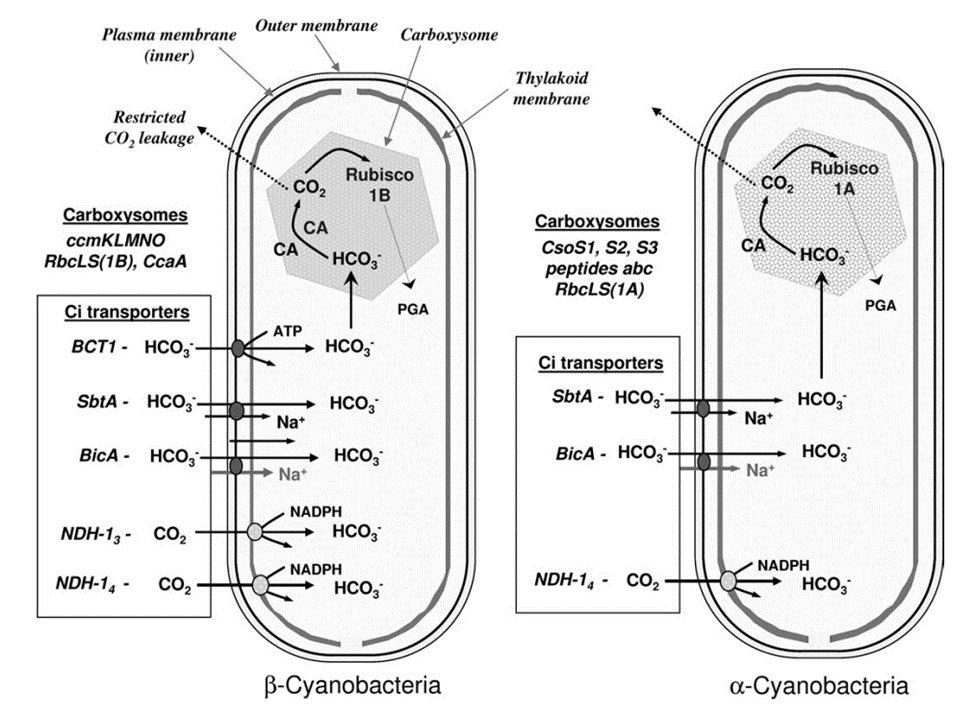
Figure 5. Characteristic components of the CO2 concentrating mechanism in α and β -cyanobacteria (Badger et al., 2006).
After HCO3– is converted to CO2, it is easier to pass the lipid layers of the cell. Thus, an effective mechanism is needed for minimizing the leakage of CO2 from the site of elevation in the carboxysome. This feature is envisaged to be associated with the shell of the carboxysome or with the molecular arrangement within the polyhedral structure: both scenarios have been theoretically modelled. The shell of the carboxysome is not a lipid membrane but a proteinaceous one. At least four different types of polypeptides have been identified that constitute this shell. In α-cyanobacteria, the proteins are CsoS1, peptides A and B. In β-cyanobacteria, the proteins are CcmK, L, and O. Together, these proteins are related to each other by containing one or more regions of homology that have become known as bacterial microcompartment domains. Especially for CcmK, this protein has been found to be able to assemble into flattened, hexamer plates of about 7 nm diameter, and may be able to assemble further into larger sheets of about 1.5 nm thick. Another interesting feature of the CcmK hexamers is that they possess a small hole (0.4 nm) in the centre of each plate that could pass small negatively charged molecules such as HCO3–, ribulose bisphosphate (RuBP), phosphoglycerate (PGA), and OH–, all of these molecules need to pass across the carboxysome coat. As a result, this proteinaceous shell is feasible to envisage a situation where charged species have superior permeability relative to uncharged species such as CO2 (Price et al., 2008).
The active uptake transporters increase the concentration of HCO3– inside the cell as a storage of Ci which is in turn used at the carboxysome to be converted into CO2 for photosynthesis. However, the level of photosynthesis is not preferred as the higher the better. The by-products of photosynthesis can cause damage to cyanobacteria cells, and the solution to this problem is explained below.
DEFENSE MECHANISMS AGAINST OXIDATIVE STRESS
Reactive Oxygen Species and Oxidative Stress
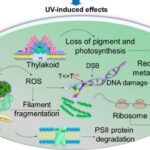
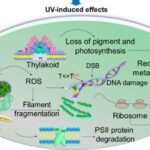
Reactive oxygen species (ROS) such as superoxide anions (O2–), hydrogen peroxide (H2O2), and hydroxyl radicals (HO•) are radical and non-radical oxygen species formed by the partial reduction of oxygen. Cellular ROS are created endogenously and generate oxidative stress that causes harmful cell damages (Ray, Huang, & Tsuji, 2012). As aerobically living organisms that also perform oxygenic photosynthesis, cyanobacteria are largely exposed to ROS produced by the respiratory machinery and the photosynthetic electron transport chain. Light, an essential component of photosynthesis, is also a major source of stress. An exposure to strong light and solar UV radiation leads to an additional production of ROS and therefore a photo-induced oxidative stress that mainly targets the photochemical efficiency of Photosystem II and inhibits this crucial step of photosynthesis (Latifi, Ruiz, & Zhang, 2009). Furthermore, oxidative stress negatively impacts pigmentation. It can also inhibit the nitrogen fixation and carbon concentrating mechanisms or alter the native protein structures and DNA of cyanobacteria (Rastogi & Madamvar, 2015). Figure 6 summarizes the harmful effects of UV-induced oxidative stress.
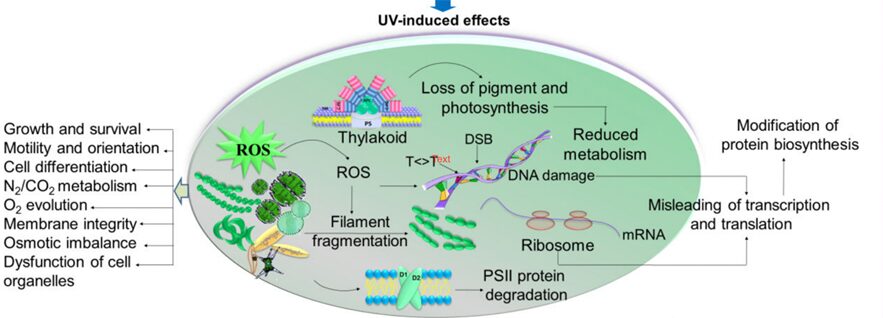
Figure 6. Summary of the UV-induced ROS production and its negative impacts on multiple aspects of cyanobacteria (Rastogi & Madamvar, 2015).
Overview of the mitigation strategies
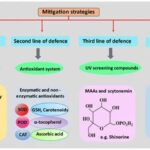
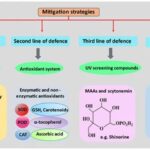
In order to perform photosynthesis and respiration, and grow in environments with high solar UV radiations, cyanobacteria have developed, in the course of their evolution, several defense mechanisms against ROS. This includes avoidance mechanisms, the production of antioxidants, the synthesis of UV absorbing compounds, and DNA repairs (Figure 7).
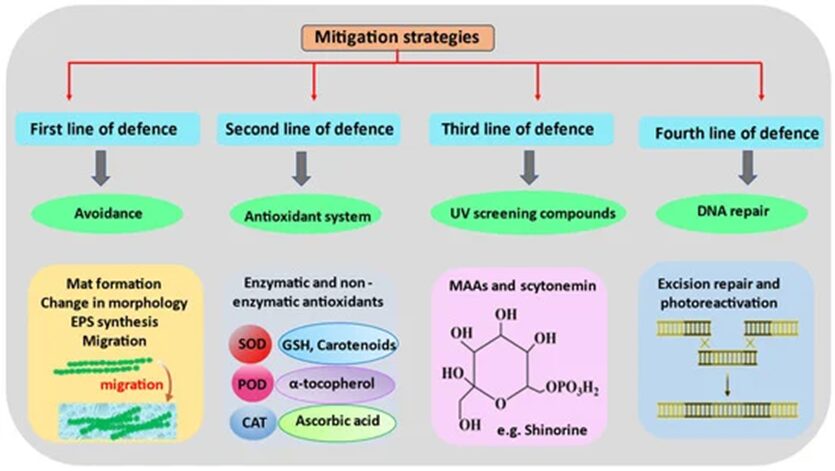
Figure 7. Chronological order of the mitigation strategies used by cyanobacteria to counteract the detrimental effects of ROS-induced stress (Singh et al., 2023).
Avoidance mechanisms
First in the line of defence are the avoidance mechanisms. In general, cyanobacteria can escape areas with high solar radiations by downward migration thanks to their gliding motility or by sinking deeper into water columns through buoyancy regulation. These two concepts are further explained in our paper focusing on the physical properties of cyanobacteria. The daily fluctuation of light intensity directs the upward or downward migration patterns of cyanobacteria in their natural habitats (Rastogi, Madamvar, & Incharoensakdi, 2015).
Besides migration, cyanobacteria also build microbial mats characterized by dense communities of microorganisms vertically stratified. This stratification is a result of the physiochemical gradients generated by the metabolic activities of the mat organisms themselves. The typical structure of a microbial mat is represented in Figure 8. Evidently, cyanobacteria need to harvest light for photosynthesis and form the top layer of microbial mats. They are covered by a layer of sand or an organic-rich mucilaginous layer containing scytonemin, a photoprotective pigment. Scytonemin is a lipid soluble, yellow-brown pigment, and naturally biosynthesized by cyanobacteria. It is capable of absorbing UV radiation and therefore protects the bacteria beneath it against UV-induced damage (Stal, 2012). The typically black or dark colours of cyanobacterial terrestrial mats is due to the presence of scytonemin (Singh et al., 2023; Stal, 2012). Furthermore, purple and green sulfur bacteria are anaerobic and perform anoxygenic photosynthesis, using sulfide as an electron donor. This means that oxygen is not released as one of the by-products. Within the mat, cyanobacteria are therefore exposed to lower oxygen concentrations, diminishing the risk of ROS formation and oxidative stress. The layer of oxidized iron serves as a barrier between the aerobic and anaerobic microbial communities. The deeper layer of the mat is black or grey due to the presence of iron sulfide (FeS) which fuels the anoxygenic photosynthesis (Stal, 2012).
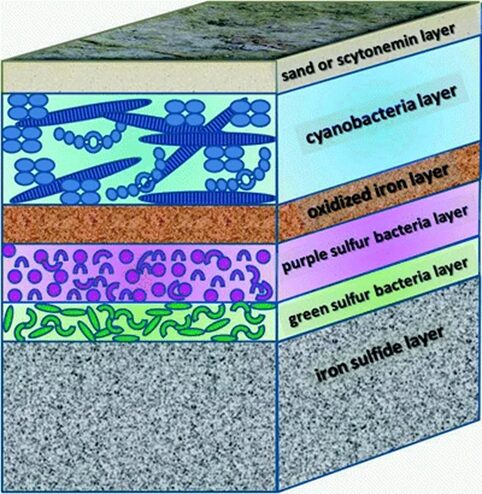
Figure 8. Schematic depicting the microbial mat formed by cyanobacteria (Stal, 2012).
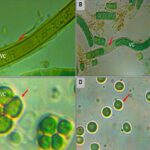
Cyanobacteria biosynthesize Extracellular Polysaccharides (EPS) as another avoidance mechanism. These high molecular weight heteropolysaccharides serve as a boundary between the cells and their surrounding environment as shown in Figure 9. Characterized by a structural diversity, EPS, depending on their thickness and consistency, play a major role in protecting cyanobacteria from UV stress. More dense EPS have a higher resistance to solar radiation and therefore a better photoprotective function. EPS provide a matrix to which scytonemin can covalently link, a significant protection against DNA strand breaks by effectively eliminating UV-induced ROS, and a free-radical-scavenging property helpful to get rid of radical ROS (Rastogi et al., 2014).
Figure 9. The EPS biosynthesized by different species of cyanobacteria (shown by the red arrows).
Antioxidant systems
Antioxidant systems are the second line of defence. As their name indicates, antioxidants significantly prevent or delay oxidation, and play a catalytic role in converting ROS and their byproducts into stable and harmless compounds making them an effective protection against oxidative stress-related cell damage (Rashi, Pankaj Kumar, & Archana, 2023). Cyanobacteria operate several antioxidant defense mechanisms to minimize the UV-induced effects caused by ROS. They consist of enzymatic systems including superoxide dismutase (SOD), catalase (CAT), peroxidase (PRX), and non-enzymatic systems involving carotenoid (CAR), and ascorbate (ASA) (Rezayian, Niknam, & Ebrahimzadeh, 2019).
Starting with antioxidant enzymes, SODs are metalloenzymes classified into four types all present in cyanobacteria: FeSOD, CuZnSOD, MnSOD, and NiSOD with the respective metal redox-active centers Fe(III), Cu(II), Zn(II), Mn(III), and Ni(II/III) used to protect cellular proteins against oxidative stress (Rashi, Pankaj Kumar, & Archana, 2023). SODs are the only enzymes capable of scavenging superoxide anions (O2–) within cells by converting them into oxygen (O2) and hydrogen peroxide (H2O2) (Singh et al., 2023). The H2O2 molecule is then detoxified by catalases or peroxidases. Catalases decompose H2O2 by binding to the hydrogen peroxide molecule at its active site and catalyzing the transfer of electrons from one H2O2 molecule to another. This results in the formation of two harmless water molecules H2O and the release of dioxygen O2 (Rastogi et al., 2014). Peroxidases are often more efficient than catalases since they can reduce hydrogen peroxides and other peroxides such as lipid hydroperoxides (ROOH) using a variety of substrates. An example is glutathione peroxidase which reduces H2O2 to water and lipid hydroperoxide to corresponding alcohols using glutathione (GHS) as an electron donor that will be oxidized to glutathione disulfide (GS-SG). Glutathione is one of the most important antioxidant enzymes for maintaining ROS homeostasis although very little is known about its roles in photosynthetic organisms (Rai et al., 2021).
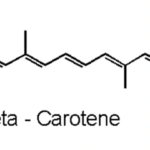
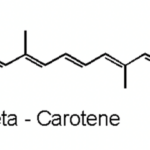
Moving on with non-enzymatic antioxidants, carotenoids are naturally occurring pigments in cyanobacteria. The polyene chain of carotenoids (Figure 10), made up of double bonds, is what gives them their coloration and their ability to absorb photons of visible wavelengths (Rashi, Pankaj Kumar, & Archana, 2023). Under prolonged UV-exposure, cyanobacteria produce a wide variety of carotenoids including but not limited to b-carotene and its derivatives. Carotenoids are localized in both the outer cellular membrane and the thylakoid membranes of cyanobacteria. They serve as light-harvesting pigments in photosynthesis and therefore are protectors of cyanobacterial cells from photooxidative harm. They achieve their protection goal by absorbing triplet-state energy from chlorophyll a, a harmful excited state ready to react with molecular oxygen to form a singlet oxygen (a strong ROS), to dissipate the additional photochemical energy and get rid of the singlet oxygen. Furthermore, in case of severe cell damage, carotenoids provide a fast SOS response, a gene activation process to repair DNA (Rastogi et al., 2014). In addition, ascorbate is a ubiquitous, less toxic, non-enzymatic antioxidant that require less energy for its synthesis. It possesses the ability to directly neutralize ROS through free radical scavenging, and to restore a-tocopherol, most commonly known as vitamin E, a remarkable antioxidant that protects Photosystem II from oxidative stress (Rai et al., 2021).
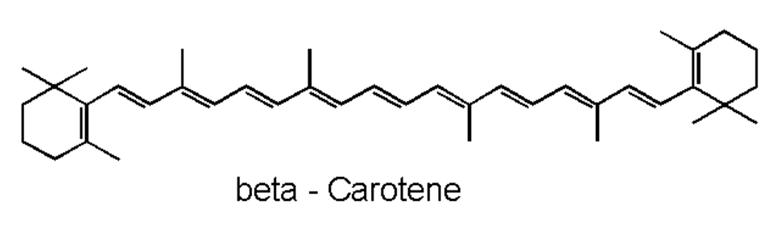
Figure 10. Representation of the polyene chain of a carotenoid, b-carotene (Carrillo, 2023).
The search for safer antioxidants derived from natural sources has raised a general interest for cyanobacterial antioxidants and their many possible applications. For example, the variety of bioactive components in cyanobacteria makes them a sustainable and feasible alternative to chemical-based fertilizers in the agricultural sector. In the field cosmetics, cyanobacteria-derived compounds are also used as natural colorants, photoprotective components in sunscreens, and antioxidant ingredients in skin products (Rashi, Pankaj Kumar, & Archana, 2023).
UV screening compounds
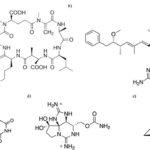
Next in the combat against oxidative stress, the biosynthesis of UV absorbing compounds by cyanobacteria is third in the line of defence. Mycosporine-like amino acids (MAAs) are the main screening compounds that maintain the organisms against UV radiation. MAAs are small, water-soluble, colourless compounds capable of absorbing all UV radiation wavelengths that reach the surface of the Earth. They vary according to the side groups attached to their central ring structure and nitrogen substituents. MAAs act as direct and indirect photoprotective compounds. By absorbing powerful UV radiation and releasing this energy as harmless heat radiation into the surroundings, MAAs act as an indirect UV-screening shield for cyanobacteria. Thanks to their antioxidant activity, MAAs are also direct free radical scavengers. They quench and interact with ROS species since the hydrogen atom from the C-4 or C-6 carbon atom is abstracted by free radicals. This result in the delocalization of the radical electron over the double bonds of the cyclohexane ring. This represents the principle of resonance shown in Figure 11. Resonance confers stability to the free radical which makes it less reactive and therefore reduces its damaging effects (Singh et al., 2023).
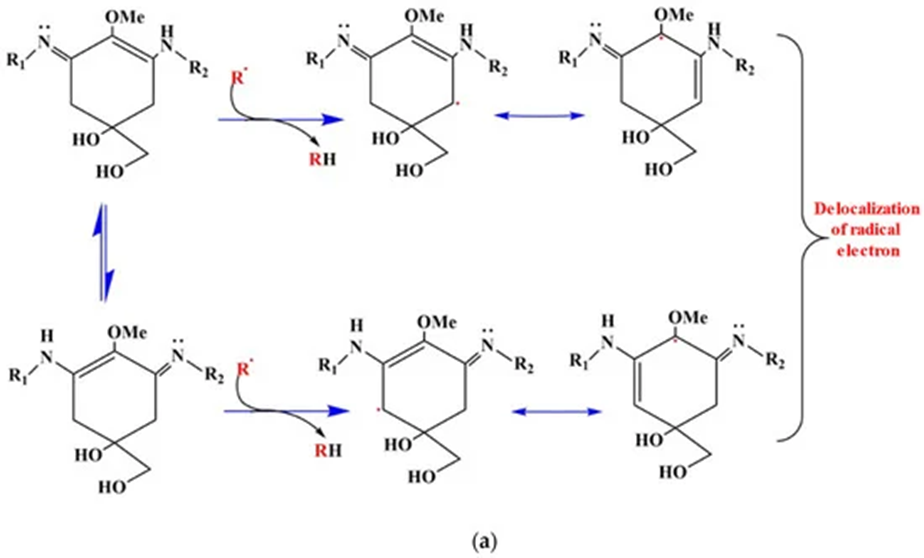
Figure 11. ROS scavenging resonance mechanism of aminocyclohexane-type MAAs. The delocalization of the radical electron leads to a more stable and less reactive configuration (Singh et al., 2023).
DNA repair mechanisms
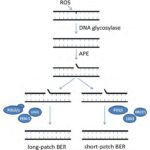
Finally, when UV-induced oxidative stress bypasses the first, second, and third lines of defence, the repair and resynthesis of complex cyanobacterial structures is the ultimate protection step. Multiple DNA repair mechanisms exist including the base excision repair and the light-dependent photoreactivation.
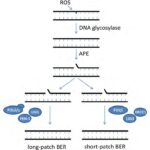
The base excision repair (BER), shown in Figure 12, is a dark pathway by which cyanobacteria repair damaged DNA during DNA replication. BER is generated by the DNA glycosylase enzymes responsible for identifying the lesions caused by ROS and removing the damaged or mutated base from the DNA helix. A nucleotide is a base linked to a sugar group and a phosphate group that participates in the formation of the backbone of DNA. DNA glycosylases cleave the N-glycosyl bond between the base and sugar and release an apurinic /apyrimidinic (AP) endonuclease enzyme. Next, this AP enzyme makes an incision at the determined base site creating a gap in the DNA strand. The strand break is then cleaned, which means that all the chemicals and intermediates present at the incision are enzymatically removed to let the repair synthesis occur. Finally, one nucleotide, in short-patch BER, or more, in long-patch BER, are synthesized to fill the break and seal the DNA strand (Corfield, 2018). This last step requires DNA polymerases beta, delta or epsilon (POLb, POLd, POLe respectively), enzymes that catalyze the resynthesis of the previously removed bases by replicating them (Wood & Shivji, 1997). It involves DNA ligases as well (LIGI, and LIGIII) which generate covalent bonds between the upstream and downstream nucleotides of the newly synthesized DNA strand (Kim & Wilson, 2012). Scaffold proteins such as XRCC1 and FEN-1 also participate in the nick-sealing process, facilitate DNA replication and promote the efficiency and speed of the repair (Izumi & Mellon, 2016).
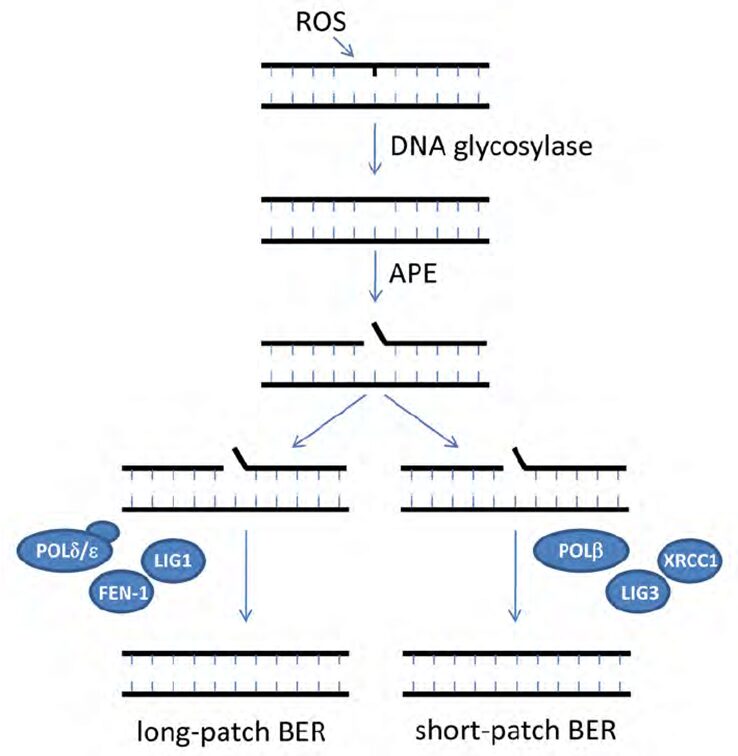
Figure 12. Schematic model for base excision repair (BER) (Biedermann, Mooney, & Hellmann, 2011).
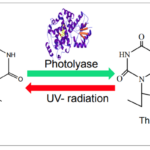
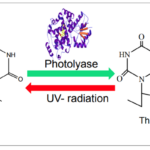
Photoreactivation by photolyase enzymes (Figure 13) is the most efficient mechanism adopted by cyanobacteria to repair cyclobutane pyrimidine dimers (CPD), the major lesion produced upon exposure of DNA to UV radiation. Light induces a covalent bond between the pyrimidine (either thymine or cytosine) bases of DNA that bind through a carbon-carbon double bond and form a dimeric structure. During photoreactivation, the bond formed between the pyrimidine bases is broken to retrieve the helix distortion of the DNA strand. Photolyases consist of flavin adenine dinucleotide (FAD) and methylene tetra hydro folate (MTHF) cofactors. The MTF absorbs blue and visible light to reach its excited state and transduce photons or electrons to FAD molecules. By accepting the photons or electrons, FAD is converted into FADH2, a higher energy molecule, and binds to the CPD to break the covalent bond. Via this photochemical reaction, photolyases reverse the CPD to the usual DNA configuration.
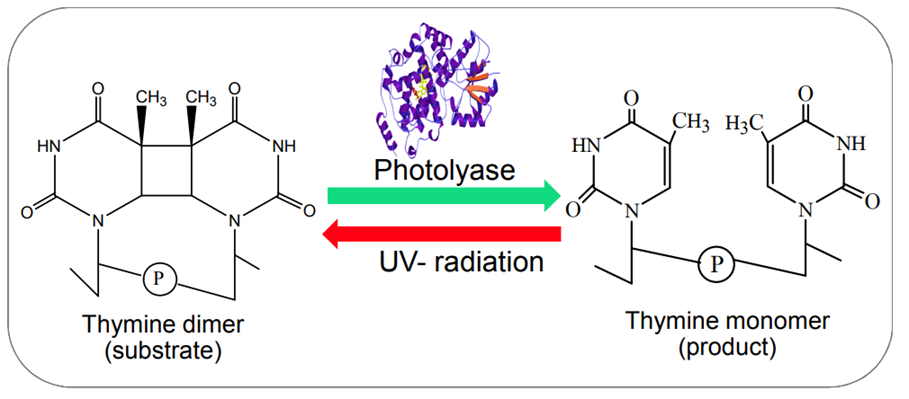
Figure 13. Example of the photoreactivation of a thymine dimer in the presence of photolyase enzymes (Rastogi & Madamvar, 2015).
In summary, cyanobacteria have developed multiple strategies to counteract the damages caused by reactive oxygen species and oxidative stress. These strategies are combined to form a line of defence starting with avoidance mechanisms, followed by antioxidants systems and the synthesis of UV-absorbing compounds, and ending with DNA repairs. Those are not the only biochemical components produced by cyanobacteria since they also release toxins to out-compete other organisms and survive in their environment.
TOXINS
Cyanobacteria produce hepatotoxins which are toxic to the liver and can cause hypovolemic shock (Vasconcelos, 2001), neurotoxins which are toxic to the nervous system, and dermatoxins which are toxic to the skin and mucous membrane (Sivonen & Jones, 1999). These toxins are secondary metabolites that can cause death and promote tumour development. Overall, toxicity is dependent on the body mass and LD50 values are a measure of the mean lethal dose of a specific substance. For example, cylindrospermopsin is a cyanotoxin, meaning a toxin synthesized by cyanobacteria, that has an LD50 value of 2.1 mg/kg at 24 hours and 0.2 mg/kg at 5-6 days, measured in mice (Sivonen & Jones, 1999). This is a cyclic guanidine alkaloid substance whose toxicity impacts the liver, heart, thymus, kidneys, and spleen by blocking protein synthesis and glutathione synthesis (Sivonen & Jones, 1999; Vasconcelos, 2001). Cyanotoxins are synthesized intracellularly and are released into water upon cell lysis or death.
Commonly produced hepatotoxins by cyanobacteria include microcystins, which are found in freshwater, and nodularin, which is found in brackish water such as the Baltic Sea (Sivonen & Jones, 1999). Both of these are cyclic peptide structures (Vasconcelos, 2001) that are tumour promotors and acts as inhibitors of serine/threonine-specific protein phosphatase (1 and 2A) (Sivonen & Jones, 1999). Protein phosphatases dephosphorylate serine and threonine residues on proteins and are essential in many physiological processes such as apoptosis, muscular contraction, protein synthesis, and glycogen metabolism (Cohen, 2010). The toxicity of hepatotoxins was studied in mice and found that they utilize bile acid carriers to infiltrate hepatocytes (Sivonen & Jones, 1999). These hepatotoxins cause hyperphosphorylation on cytoskeleton proteins leading to cell shrinkage (Vasconcelos, 2001), subsequent internal hemorrhage, and shock (Sivonen & Jones, 1999). Hepatocytes are the essential cells in the liver that are involved in metabolism and detoxification (Zhou, Xu, & Gao, 2016). They also release proteins into the bloodstream that are critical for immune responses such as targeting and annihilating pathogens or facilitating phagocytosis.
Microcystis, Planktothrix, and Anabaena are cyanobacteria genera that create microcystin (Sivonen & Jones, 1999). It has been noted throughout the cell’s growth and development, but production particularly increases when the cyanobacteria are in favourable growth conditions. Its structure is “cyclo(D-alanine1 -X2 -D-MeAsp3 -Z4 – Adda5 -D-glutamate6 -Mdha7), in which X and Z are variable L-amino acids, D-MeAsp3 is D-erythro-ß-methyl aspartic acid, and Mdha is N-methyldehydroalanine” (Sivonen & Jones, 1999). Knockout genomics is the process of inactivating genes to study their functions as related to an organism. This type of genetic engineering has been utilized to understand gene clusters involved in the synthesis of microcystin in Microcystis aeruginosa PCC 7806 and K-39 species (Sivonen & Jones, 1999). Adda, an amino acid in microcystin, is encoded by mcyD, mcyG, and mcyE genes and the synthesis pentapeptide component of microcystin is governed by mcyA, mcyB, and mcyc genes. Nodularin is produced by Nodularia spumigena and its structure is cyclo(D-MeAsp1 -L-arginine2 -Adda3 -Dglutamate4 -Mdhb5), in which Mdhb is 2-(methylamino)- 2-dehydrobutyric acid. Microcystin and nodularin production also involves non-ribosomal peptide synthases. Non-ribosomal peptide synthases adopt “a step-by-step mechanism where a single module is responsible for the activation, thioester formation, condensation and optionally modification of a single amino acid unit” (Pearson et al., 2016).
Anatoxin-a, anatoxin-a(S), and saxitoxins are neurotoxins produced by various cyanobacteria and have been reported to cause respiratory arrest in animals (Sivonen & Jones, 1999; Vasconcelos, 2001). Firstly, anatoxin-a is an alkaloid with a structure of 2-acetyl-9-azabicyclo(4-2-1)non-2-ene as shown in Figure 14 and functions similarly to the neurochemical acetylcholine which is responsible notably for muscular contraction, memory and attention. Since it can also bind to acetylcholine receptors, but has a different chemical composition, it cannot be broken down by acetylcholinesterase and therefore will continue to stimulate cholinergic nerves (Vasconcelos, 2001). This can result in paralysis. It can be produced by Oscillatoria, Planktothrix rubescens, Phormidium flavosum (Sivonen & Jones, 1999). Anatoxin-a(S) is an anticholinesterase which inhibits the enzymes from breaking down acetylcholine resulting in excessive stimulation. This is another toxin that can induce paralysis (Vasconcelos, 2001). It is produced by Anabaena flos-aquae strain NRC 525-17 and its structure is a phosphaste ester cyclic N-hydroxyguanine (Sivonen & Jones, 1999). Lastly, saxitoxins function by blocking sodium channels on nerve cell membranes and on skeletal muscle cells (Vasconcelos, 2001). These sodium channels are critical for signal propagation and generation, thus when the diffusion of sodium ions is inhibited, it can lead to fatality. They can be synthesized by many cyanobacteria such as Anabaena circinalis, Cylindrospermopsis raciborskii, and Lyngbya wollei, and are carbamate alkaloids (Sivonen & Jones, 1999). Saxitoxin is synthesized by a methylation of acytyl-CoA, an arginine condensation, an addition of an amidino group with the help of aminotransferase SxtG, followed by the creation of tricyclic core. This occurs through cyclization and desaturation reactions. The enzyme carbomoyltransferase lastly adds a carbomyl substituent (Pearson et al., 2016).
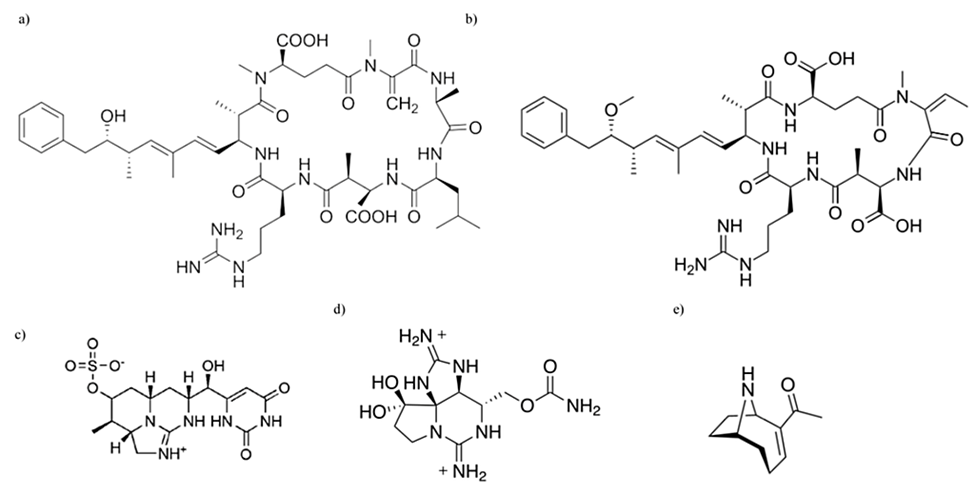
Figure 14. Chemical structural differences between various cyanotoxins: (a) microcystin, (b) nodularin, (c) cylindrospermopsin, (d) saxitoxin, (e) anatoxin-a (Holland & Kinnear, 2013).
Bioactive Agents
Cyanobacteria also produce bioactive agents, meaning substances that can alter tissue, cell, or organism functions. Curacin A is such an example, disrupting mitosis and supressing cell growth, and it is produced by multienzyme complexes in Lyngbya sp cells (Sivonen & Jones, 1999). Nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) encode these complexes. Barbamide and jamaicamides are also bioactive substances produced by this species with the former acting as mullusk pesticide and the latter inhibiting sodium channel activity. However, not all bioactive agents produced by cyanobacteria are inherently dangerous or toxic to humans. In fact, some even have antiviral properties which have been studied for our future use. Cyanovirin-N produced by Nostoc ellipsosporum is effective at inhibiting the activity of human immunodeficiency virus (HIV) “by bind[ing] to N-linked high-mannose oligosaccharides on the viral envelope” (Sivonen & Jones, 1999).
Advantages of Toxin Production
It is contested whether cyanobacteria toxins are significantly advantageous by giving a cellular defense mechanism or by the amelioration of its physiological processes. For example, some studies report that exposure to cyanotoxins inhibits Diatom growth, reproductive capabilities of Cryptomonas (Vasconcelos, 2001), and the feeding rate of Daphnia pulicaria (Holland & Kinnear, 2013). As well, microcystin and anatoxin-a have the ability to inhibit the movement of Chlamydomonas reinhardtii. However, it has also been reported that the inhibition of growth of other species, such as phytoplankton, is not significant. As well, concentrations of cyanotoxins exceeding those present in the environment has been shown to cause acute toxicity in aquatic plants like Spirodela oligorrhiza, but these results are not representative of normal interactions. It is proposed that these toxins may be physiologically advantageous to cyanobacteria. Saxitoxin can block sodium channels to maintain homeostasis when its concentration causes sodium stress. Additionally, phytoplankton in response to exposure of cylindrospermopsin release alkaline phosphatase and cyanobacteria benefit from increased availability of inorganic phosphate. These toxins have also been used as signalling chemicals to other cyanobacteria, such that upon cell rupture and exposure of cyanotoxins such as microcystin, other cyanobacteria increased their production of those substances. This may reduce the palatability of these organisms as prey.
CONCLUSION
In conclusion, cyanobacteria are living biochemical factories. They make a great use of their chemical properties and have developed unique mechanisms to survive under harsh conditions, to combat environmental stresses, and to out-compete other organisms.
Indeed, to adapt to difficult environmental conditions or nutrient-poor environments for photosynthesis, nature has equipped cyanobacteria with nitrogenases, enzymes that allow them to fix nitrogen which can be used in all living organisms. Furthermore, to protect these nitrogenases against O2 degradation, cyanobacteria employ strategies that include spatial separation, fixing nitrogen at night, and some cyanobacteria even employ a combination of these techniques. The ability to fix nitrogen has facilitated the formation of numerous symbiotic relationships in which cyanobacteria benefit from enhanced carbon availability, protection, and ideal chemical environments for growth and prosperity.
Despite being under CO2 limiting conditions, the Rubisco enzymes of cyanobacteria have a low affinity to CO2 which seems to make the problem of limited photosynthesis worse. However, cyanobacteria are still able to achieve a saturated rate of CO2 fixation by actively uptaking and storing a high concentration of HCO3– in carboxysomes.
Furthermore, by performing photosynthesis and aerobic respiration, and by being exposed to UV radiation for long periods of time, cyanobacteria often come across reactive oxygen species (ROS). To combat the ROS-induced oxidative stress, cyanobacteria have evolved a series of defense mechanisms. These mitigation strategies involve avoidance mechanisms such as migration, the formation of microbial mats, and the biosynthesis of extracellular polysaccharides. Additionally, cyanobacteria employ antioxidant enzymatic and non-enzymatic systems to neutralize ROS. They also synthesize UV-absorbing compounds known as mycosporine-like amino acids and have DNA repair mechanisms including base excision repair and photoreactivation by photolyase to limit or fix damages caused by a prolonged exposure to UV radiation.
Last but not least, to out-compete other organisms for nutrients and optimal habitat, as well support physiological homeostasis, cyanobacteria have adapted to produce toxins and other bioactive agents. Some of these block sodium channels on cell membranes to regulate its own concentration intracellularly during sodium stress, while also functioning to inhibit other organisms’ motility upon release. Microcystins notably act as toxins by inhibiting serine/threonine-specific protein phosphatase which leads to hyperphosphorylated proteins and can cause cytoskeleton deformation and cell shrinkage. Neurotoxins like anatoxin-a which acts like acetylcholine and anatoxin-a(s) which is an anticholinesterase, cause continuous stimulation of cholinergic nerves in other organisms leading to paralysis and even death. Other toxins can inhibit the reproduction and feeding rate of other organisms, or even cause phytoplankton to release inorganic phosphate to cyanobacteria’s advantage.
Overall, cyanobacteria are full of impressive chemical properties. Understanding them is vital for various fields such as environmental sciences, biotechnology, pharmaceuticals, agriculture, cosmetics, and many more. It is crucial to harness their potential benefits and mitigate their possible harms.
References
Badger, M. R., Price, G. D., Long, B. M., & Woodger, F. J. (2006). The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. Journal of experimental botany, 57(2), 249-265.
Berman-Frank, I., Lundgren, P., & Falkowski, P. (2003). Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Research in Microbiology, 154(3), 157-164. https://doi.org/https://doi.org/10.1016/S0923-2508(03)00029-9
Biedermann, S., Mooney, S., & Hellmann, H. (2011). Recognition and Repair Pathways of Damaged DNA in Higher Plants. In. https://doi.org/10.5772/21380
Björn, W., & Karl, F. (2018). Cyanophycin: A Nitrogen-Rich Reserve Polymer. In T. Archana (Ed.), Cyanobacteria (pp. Ch. 5). IntechOpen. https://doi.org/10.5772/intechopen.77049
Bothe, H., Schmitz, O., Yates, M. G., Newton, W. E. . (2010). Nitrogen Fixation and Hydrogen Metabolism in Cyanobacteria. Microbiology and Molecular Biology Reviews, 74(4), 529–551. https://doi.org/10.1128/mmbr.00033-10
Britannica. (2023). nitrogen fixation. Encyclopedia Britannica. Retrieved November 4 from https://www.britannica.com/science/nitrogen-fixation
Carrillo, J.-C. (2023). Oxidants and Antioxidants in Human Health. Nutritional Intervention Study Based on the Campaign “5 a Day”.
Cohen, P. (2010). Chapter 85-Phosphatase Families Dephosphorylating Serine and Threonine Residues in Proteins A2-Bradshaw, Ralph A. In: Handbook of Cell Signaling (Second Edition). San Diego: Academic Press.
Corfield, J. (2018). Base Excision Repair. Encyclopedia Britannica. https://www.britannica.com/science/base-excision-repair
Fujita, Y., & Uesaka, K. (2022). Chapter 3 – Nitrogen fixation in cyanobacteria. In H. Kageyama & R. Waditee-Sirisattha (Eds.), Cyanobacterial Physiology (pp. 29-45). Academic Press. https://doi.org/https://doi.org/10.1016/B978-0-323-96106-6.00007-1
Holland, A., & Kinnear, S. (2013). Interpreting the possible ecological role (s) of cyanotoxins: compounds for competitive advantage and/or physiological aide? Marine drugs, 11(7), 2239-2258.
Izumi, T., & Mellon, I. (2016). Chapter 17 – Base Excision Repair and Nucleotide Excision Repair. In I. Kovalchuk & O. Kovalchuk (Eds.), Genome Stability (pp. 275-302). Academic Press. https://doi.org/https://doi.org/10.1016/B978-0-12-803309-8.00017-3
Jabbur, M. L., & Johnson, C. H. (2021). Spectres of Clock Evolution: Past, Present, and Yet to Come. Front Physiol, 12, 815847. https://doi.org/10.3389/fphys.2021.815847
Jansson, C., & Northen, T. (2010). Calcifying cyanobacteria—the potential of biomineralization for carbon capture and storage. Current opinion in biotechnology, 21(3), 365-371.
Kamennaya, N. A., Ajo-Franklin, C. M., Northen, T., & Jansson, C. (2012). Cyanobacteria as biocatalysts for carbonate mineralization. Minerals, 2(4), 338-364.
Kim, Y. J., & Wilson, D. M., 3rd. (2012). Overview of base excision repair biochemistry. Curr Mol Pharmacol, 5(1), 3-13. https://doi.org/10.2174/1874467211205010003
Kumar, K., Mella-Herrera, R. A., & Golden, J. W. (2010). Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol, 2(4), a000315. https://doi.org/10.1101/cshperspect.a000315
Latifi, A., Ruiz, M., & Zhang, C.-C. (2009). Oxidative stress in cyanobacteria. FEMS Microbiology Reviews, 33(2), 258-278. https://doi.org/10.1111/j.1574-6976.2008.00134.x
Latysheva, N., Junker, V. L., Palmer, W. J., Codd, G. A., & Barker, D. (2012). The evolution of nitrogen fixation in cyanobacteria. Bioinformatics, 28(5), 603-606. https://doi.org/10.1093/bioinformatics/bts008
Lesser, M. P., Mazel, C. H., Gorbunov, M. Y., & Falkowski, P. G. (2004). Discovery of Symbiotic Nitrogen-Fixing Cyanobacteria in Corals. Science, 305(5686), 997-1000. https://doi.org/doi:10.1126/science.1099128
Meeks, J. C. (1998). Symbiosis between Nitrogen-Fixing Cyanobacteria and Plants. BioScience, 48(4), 266-276. https://doi.org/10.2307/1313353
Mehdizadeh Allaf, M., & Peerhossaini, H. (2022). Cyanobacteria: Model Microorganisms and Beyond. Microorganisms, 10(4). https://doi.org/10.3390/microorganisms10040696
Pearson, L. A., Dittmann, E., Mazmouz, R., Ongley, S. E., D’Agostino, P. M., & Neilan, B. A. (2016). The genetics, biosynthesis and regulation of toxic specialized metabolites of cyanobacteria. Harmful Algae, 54, 98-111.
Price, G. D., Badger, M. R., Woodger, F. J., & Long, B. M. (2008). Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. Journal of experimental botany, 59(7), 1441-1461.
Rai, R., Singh, S., Rai, K. K., Raj, A., Sriwastaw, S., & Rai, L. C. (2021). Regulation of antioxidant defense and glyoxalase systems in cyanobacteria. Plant Physiology and Biochemistry, 168, 353-372. https://doi.org/https://doi.org/10.1016/j.plaphy.2021.09.037
Rashi, T., Pankaj Kumar, S., & Archana, T. (2023). Cyanobacteria as the Source of Antioxidants. In T. Archana (Ed.), Cyanobacteria (pp. Ch. 3). IntechOpen. https://doi.org/10.5772/intechopen.110598
Rastogi, R., & Madamvar, D. (2015). UV-Induced Oxidative Stress in Cyanobacteria: How Life is able to Survive? Biochemistry & Analytical Biochemistry, 4. https://doi.org/10.4172/2161-1009.1000173
Rastogi, R., Madamvar, D., & Incharoensakdi, A. (2015). Multiple defense systems in cyanobacteria in response to solar UV radiation. In (pp. 125-158). https://doi.org/10.13140/RG.2.1.3270.0889
Rastogi, R. P., Sinha, R. P., Moh, S. H., Lee, T. K., Kottuparambil, S., Kim, Y.-J., Rhee, J.-S., Choi, E.-M., Brown, M. T., Häder, D.-P., & Han, T. (2014). Ultraviolet radiation and cyanobacteria. Journal of Photochemistry and Photobiology B: Biology, 141, 154-169. https://doi.org/https://doi.org/10.1016/j.jphotobiol.2014.09.020
Ray, P. D., Huang, B. W., & Tsuji, Y. (2012). Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal, 24(5), 981-990. https://doi.org/10.1016/j.cellsig.2012.01.008
Rezayian, M., Niknam, V., & Ebrahimzadeh, H. (2019). Stress response in cyanobacteria. Iranian Journal of Plant Physiology, 9(3), 2773-2787. https://doi.org/10.30495/ijpp.2019.666767
Sánchez-Baracaldo, P., & Cardona, T. (2020). On the origin of oxygenic photosynthesis and Cyanobacteria. New Phytologist, 225(4), 1440-1446. https://doi.org/https://doi.org/10.1111/nph.16249
Singh, V. K., Jha, S., Rana, P., Mishra, S., Kumari, N., Singh, S. C., Anand, S., Upadhye, V., & Sinha, R. P. (2023). Resilience and Mitigation Strategies of Cyanobacteria under Ultraviolet Radiation Stress. International Journal of Molecular Sciences, 24(15), 12381. https://www.mdpi.com/1422-0067/24/15/12381
Sivonen, K., & Jones, G. (1999). Cyanobacterial toxins. Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management, 1, 43-112.
Stal, L. J. (2012). Cyanobacterial Mats and Stromatolites. In B. A. Whitton (Ed.), Ecology of Cyanobacteria II: Their Diversity in Space and Time (pp. 65-125). Springer Netherlands. https://doi.org/10.1007/978-94-007-3855-3_4
Stal, L. J. (2015). Nitrogen Fixation in Cyanobacteria. In Encyclopedia of Life Sciences (pp. 1-9). https://doi.org/https://doi.org/10.1002/9780470015902.a0021159.pub2
Strohmeyer, C. (2019, 1/22/2019). Cyanobacteria; Blue Green/Red Slime Algae in Aquariums & Ponds. Retrieved 11/15/2023 from
Vasconcelos, V. (2001). Cyanobacteria toxins: diversity and ecological effects. Limnetica, 20(1), 45-58.
Vermaas, W. (2001). Photosynthesis and Respiration in Cyanobacteria. In (Vol. 1–7). https://doi.org/10.1038/npg.els.0001670
Wood, R. D., & Shivji, M. K. (1997). Which DNA polymerases are used for DNA-repair in eukaryotes? Carcinogenesis, 18(4), 605-610. https://doi.org/10.1093/carcin/18.4.605
Zhou, Z., Xu, M.-J., & Gao, B. (2016). Hepatocytes: a key cell type for innate immunity. Cellular & molecular immunology, 13(3), 301-315.